Hello,
I am trying to calucate the mean of one of my columns "response" but it seems to come up with NH when I try to calculate it. Here
below is what I have typed in so far:library(tidyverse)
library(tidyverse)
dat <- read_csv("Learningtask.csv")
tidydat <- dat %>% select(participant, response_age_a,response_age_b, response_gender, stage, trial_type, cue, correct_response, response, correct, rt, block_s1, count_s1, count_s2)
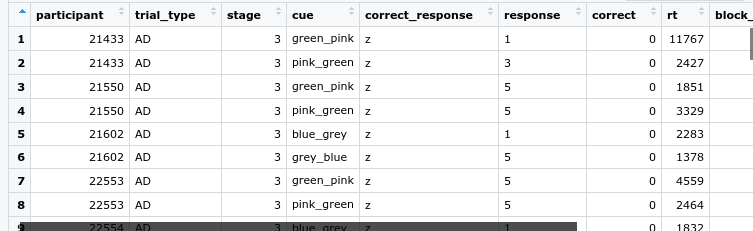
dat %>% head ()
dat%>% glimpse()
dat_clean <-
dat %>% select(participant,
trial_type,
stage,
cue,
correct_response,
response,
correct,
rt,
block_s1,
count_s1,
count_s2)
colnames(dat_clean) <- c("participant",
"trial_type",
"stage",
"cue",
"correct_response",
"response",
"correct",
"rt",
"block_s1",
"count_s1",
"count_s2")
exclude <- c(22617, 22638, 21470, 22557, 22666, 22701, 22714, 22736, 22759, 22806,23180, 23218, 23271, 23273, 23300, 23341, 23465, 23469, 22617,22638, 21470, 22557, 22736, 22759, 23218, 23341, 23465)
dat_clean <- dat_clean %>% filter(!(participant %in% exclude))
head(dat_clean)
nc_include2 <- dat_clean %>% filter(stage =="3") %>% drop_na()
nc_includead <- nc_include2 %>% filter (trial_type == "AD")
Once I have filtered out my data I type in this sentence:
nc_includead %>% summarise(mean(response))
and it comes up with this in the output:
nc_includead %>% summarise(mean(response))
A tibble: 1 x 1
mean(response)
1 NA
Warning message:
In mean.default(response) :
argument is not numeric or logical: returning NA
I was wondering if anyone could me understand why this is happening and how to fix it.