This is a companion discussion topic for the original entry at https://blogs.rstudio.com/tensorflow/posts/2021-04-22-torch-for-optimization

Photo by Michael Trimble on Unsplash
So far, all torch use cases we’ve discussed here have been in deep learning. However, its automatic differentiation feature is useful in other areas. One prominent example is numerical optimization: We can use torch to find the minimum of a function.
In fact, function minimization is exactly what happens in training a neural network. But there, the function in question normally is far too complex to even imagine finding its minima analytically. Numerical optimization aims at building up the tools to handle just this complexity. To that end, however, it starts from functions that are far less deeply composed. Instead, they are hand-crafted to pose specific challenges.
This post is a first introduction to numerical optimization with torch . Central takeaways are the existence and usefulness of its L-BFGS optimizer, as well as the impact of running L-BFGS with line search. As a fun add-on, we show an example of constrained optimization, where a constraint is enforced via a quadratic penalty function.
To warm up, we take a detour, minimizing a function “ourselves” using nothing but tensors. This will turn out to be relevant later, though, as the overall process will still be the same. All changes will be related to integration of optimizer s and their capabilities.
Function minimization, DYI approach
To see how we can minimize a function “by hand”, let’s try the iconic Rosenbrock function. This is a function with two variables:
, with aa and bb configurable parameters often set to 1 and 5, respectively.
In R:
library(torch)
a <- 1
b <- 5
rosenbrock <- function(x) {
x1 <- x[1]
x2 <- x[2]
(a - x1)^2 + b * (x2 - x1^2)^2
}
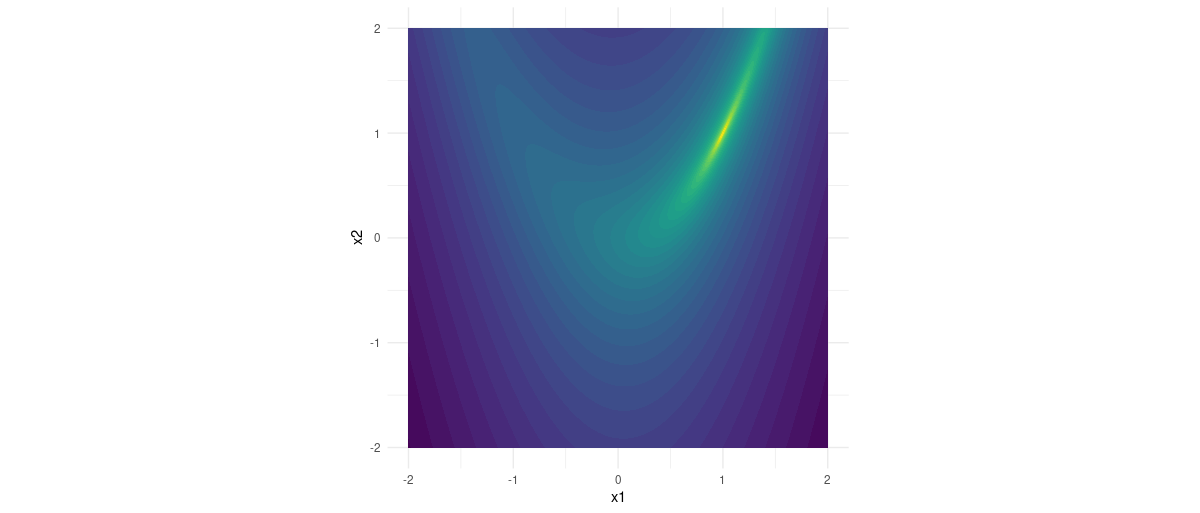
Its minimum is located at (1,1), inside a narrow valley surrounded by breakneck-steep cliffs: 1
Figure 1: Rosenbrock function.
Our goal and strategy are as follows.
We want to find the values x1 and x2 for which the function attains its minimum. We have to start somewhere; and from wherever that gets us on the graph we follow the negative of the gradient “downwards”, descending into regions of consecutively smaller function value.
Concretely, in every iteration, we take the current (x_1,x_2) point, compute the function value as well as the gradient, and subtract some fraction of the latter to arrive at a new (x_1,x_2) candidate. This process goes on until we either reach the minimum – the gradient is zero – or improvement is below a chosen threshold.
Here is the corresponding code. For no special reasons, we start at (-1,1) . The learning rate (the fraction of the gradient to subtract) needs some experimentation. (Try 0.1 and 0.001 to see its impact.)
num_iterations <- 1000
# fraction of the gradient to subtract
lr <- 0.01
# function input (x1,x2)
# this is the tensor w.r.t. which we'll have torch compute the gradient
x_star <- torch_tensor(c(-1, 1), requires_grad = TRUE)
for (i in 1:num_iterations) {
if (i %% 100 == 0) cat("Iteration: ", i, "\n")
# call function
value <- rosenbrock(x_star)
if (i %% 100 == 0) cat("Value is: ", as.numeric(value), "\n")
# compute gradient of value w.r.t. params
value$backward()
if (i %% 100 == 0) cat("Gradient is: ", as.matrix(x_star$grad), "\n\n")
# manual update
with_no_grad({
x_star$sub_(lr * x_star$grad)
x_star$grad$zero_()
})
}
Iteration: 100
Value is: 0.3502924
Gradient is: -0.667685 -0.5771312
Iteration: 200
Value is: 0.07398106
Gradient is: -0.1603189 -0.2532476
...
...
Iteration: 900
Value is: 0.0001532408
Gradient is: -0.004811743 -0.009894371
Iteration: 1000
Value is: 6.962555e-05
Gradient is: -0.003222887 -0.006653666
While this works, it really serves to illustrate the principle. With torch providing a bunch of proven optimization algorithms, there is no need for us to manually compute the candidate xx values.
Function minimization with torch optimizers
Instead, we let a torch optimizer update the candidate xx for us. Habitually, our first try is Adam .
Adam
With Adam, optimization proceeds a lot faster. Truth be told, though, choosing a good learning rate still takes non-negligeable experimentation. (Try the default learning rate, 0.001, for comparison.)
num_iterations <- 100
x_star <- torch_tensor(c(-1, 1), requires_grad = TRUE)
lr <- 1
optimizer <- optim_adam(x_star, lr)
for (i in 1:num_iterations) {
if (i %% 10 == 0) cat("Iteration: ", i, "\n")
optimizer$zero_grad()
value <- rosenbrock(x_star)
if (i %% 10 == 0) cat("Value is: ", as.numeric(value), "\n")
value$backward()
optimizer$step()
if (i %% 10 == 0) cat("Gradient is: ", as.matrix(x_star$grad), "\n\n")
}
Iteration: 10
Value is: 0.8559565
Gradient is: -1.732036 -0.5898831
Iteration: 20
Value is: 0.1282992
Gradient is: -3.22681 1.577383
...
...
Iteration: 90
Value is: 4.003079e-05
Gradient is: -0.05383469 0.02346456
Iteration: 100
Value is: 6.937736e-05
Gradient is: -0.003240437 -0.006630421
It took us about a hundred iterations to arrive at a decent value. This is a lot faster than the manual approach above, but still quite a lot. Luckily, further improvements are possible.
L-BFGS
Among the many torch optimizers commonly used in deep learning (Adam, AdamW, RMSprop …), there is one “outsider”, much better known in classic numerical optimization than in neural-networks space: L-BFGS, a.k.a. Limited-memory BFGS, a memory-optimized implementation of the Broyden–Fletcher–Goldfarb–Shanno optimization algorithm (BFGS).
BFGS is perhaps the most widely used among the so-called Quasi-Newton, second-order optimization algorithms. As opposed to the family of first-order algorithms that, in deciding on a descent direction, make use of gradient information only, second-order algorithms additionally take curvature information into account. To that end, exact Newton methods actually compute the Hessian (a costly operation), while Quasi-Newton methods avoid that cost and, instead, resort to iterative approximation.
Looking at the contours of the Rosenbrock function, with its prolonged, narrow valley, it is not difficult to imagine that curvature information might make a difference. And, as you’ll see in a second, it really does. Before though, one note on the code. When using L-BFGS, it is necessary to wrap both function call and gradient evaluation in a closure ( calc_loss() , in the below snippet), for them to be callable several times per iteration. You can convince yourself that the closure is, in fact, entered repeatedly, by inspecting this code snippet’s chatty output:
num_iterations <- 3
x_star <- torch_tensor(c(-1, 1), requires_grad = TRUE)
optimizer <- optim_lbfgs(x_star)
calc_loss <- function() {
optimizer$zero_grad()
value <- rosenbrock(x_star)
cat("Value is: ", as.numeric(value), "\n")
value$backward()
cat("Gradient is: ", as.matrix(x_star$grad), "\n\n")
value
}
for (i in 1:num_iterations) {
cat("Iteration: ", i, "\n")
optimizer$step(calc_loss)
}
Iteration: 1
Value is: 4
Gradient is: -4 0
Value is: 6
Gradient is: -2 10
...
...
Value is: 0.04880721
Gradient is: -0.262119 -0.1132655
Value is: 0.0302862
Gradient is: 1.293824 -0.7403332
Iteration: 2
Value is: 0.01697086
Gradient is: 0.3468466 -0.3173429
Value is: 0.01124081
Gradient is: 0.2420997 -0.2347881
...
...
Value is: 1.111701e-09
Gradient is: 0.0002865837 -0.0001251698
Value is: 4.547474e-12
Gradient is: -1.907349e-05 9.536743e-06
Iteration: 3
Value is: 4.547474e-12
Gradient is: -1.907349e-05 9.536743e-06
Even though we ran the algorithm for three iterations, the optimal value really is reached after two. Seeing how well this worked, we try L-BFGS on a more difficult function, named flower , for pretty self-evident reasons.
(Yet) more fun with L-BFGS
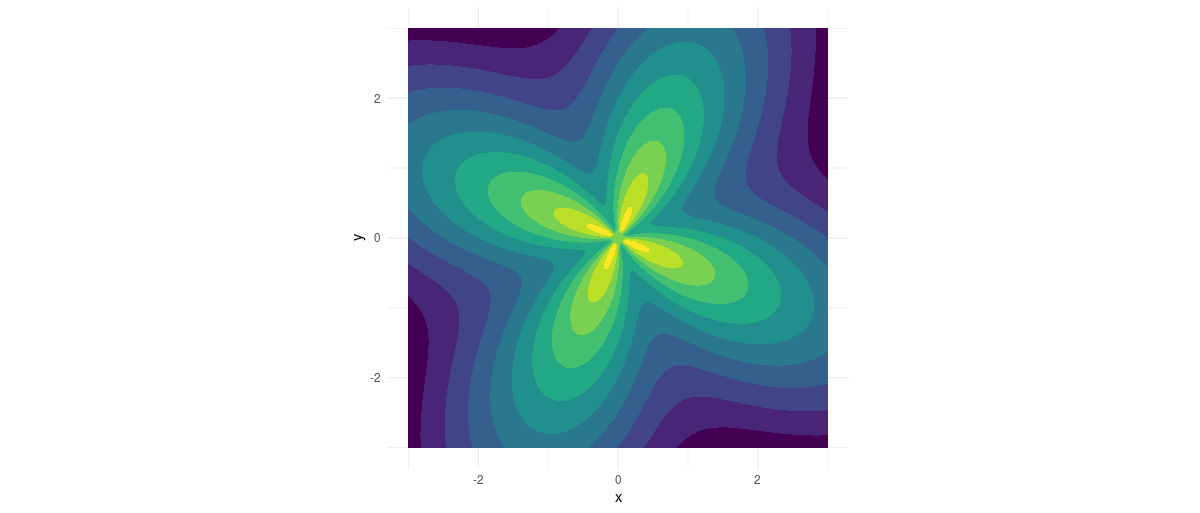
Here is the flower function. 2 Mathematically, its minimum is near (0,0) , but technically the function itself is undefined at (0,0) , since the atan2 used in the function is not defined there.
a <- 1
b <- 1
c <- 4
flower <- function(x) {
a * torch_norm(x) + b * torch_sin(c * torch_atan2(x[2], x[1]))
}
Figure 2: Flower function.
We run the same code as above, starting from (20,20) this time.
num_iterations <- 3
x_star <- torch_tensor(c(20, 0), requires_grad = TRUE)
optimizer <- optim_lbfgs(x_star)
calc_loss <- function() {
optimizer$zero_grad()
value <- flower(x_star)
cat("Value is: ", as.numeric(value), "\n")
value$backward()
cat("Gradient is: ", as.matrix(x_star$grad), "\n")
cat("X is: ", as.matrix(x_star), "\n\n")
value
}
for (i in 1:num_iterations) {
cat("Iteration: ", i, "\n")
optimizer$step(calc_loss)
}
Iteration: 1
Value is: 28.28427
Gradient is: 0.8071069 0.6071068
X is: 20 20
...
...
Value is: 19.33546
Gradient is: 0.8100872 0.6188223
X is: 12.957 14.68274
...
...
Value is: 18.29546
Gradient is: 0.8096464 0.622064
X is: 12.14691 14.06392
...
...
Value is: 9.853705
Gradient is: 0.7546976 0.7025688
X is: 5.763702 8.895616
Value is: 2635.866
Gradient is: -0.7407354 -0.6717985
X is: -1949.697 -1773.551
Iteration: 2
Value is: 1333.113
Gradient is: -0.7413024 -0.6711776
X is: -985.4553 -897.5367
Value is: 30.16862
Gradient is: -0.7903821 -0.6266789
X is: -21.02814 -21.72296
Value is: 1281.39
Gradient is: 0.7544561 0.6563575
X is: 964.0121 843.7817
Value is: 628.1306
Gradient is: 0.7616636 0.6480014
X is: 475.7051 409.7372
Value is: 4965690
Gradient is: -0.7493951 -0.662123
X is: -3721262 -3287901
Value is: 2482306
Gradient is: -0.7503822 -0.6610042
X is: -1862675 -1640817
Value is: 8.61863e+11
Gradient is: 0.7486113 0.6630091
X is: 645200412672 571423064064
Value is: 430929412096
Gradient is: 0.7487153 0.6628917
X is: 322643460096 285659529216
Value is: Inf
Gradient is: 0 0
X is: -2.826342e+19 -2.503904e+19
Iteration: 3
Value is: Inf
Gradient is: 0 0
X is: -2.826342e+19 -2.503904e+19
This has been less of a success. At first, loss decreases nicely, but suddenly, the estimate dramatically overshoots, and keeps bouncing between negative and positive outer space ever after.
Luckily, there is something we can do.
L-BFGS with line search
Taken in isolation, what a Quasi-Newton method like L-BFGS does is determine the best descent direction. However, as we just saw, a good direction is not enough. With the flower function, wherever we are, the optimal path leads to disaster if we stay on it long enough. Thus, we need an algorithm that carefully evaluates not only where to go, but also, how far.
For this reason, L-BFGS implementations commonly incorporate line search , that is, a set of rules indicating whether a proposed step length is a good one, or should be improved upon.
Specifically, torch ’s L-BFGS optimizer implements the Strong Wolfe conditions. We re-run the above code, changing just two lines. Most importantly, the one where the optimizer is instantiated:
optimizer <- optim_lbfgs(x_star, line_search_fn = "strong_wolfe")
And secondly, this time I found that after the third iteration, loss continued to decrease for a while, so I let it run for five iterations. Here is the output:
Iteration: 1
...
...
Value is: -0.8838741
Gradient is: 3.742207 7.521572
X is: 0.09035123 -0.03220009
Value is: -0.928809
Gradient is: 1.464702 0.9466625
X is: 0.06564617 -0.026706
Iteration: 2
...
...
Value is: -0.9991404
Gradient is: 39.28394 93.40318
X is: 0.0006493925 -0.0002656128
Value is: -0.9992246
Gradient is: 6.372203 12.79636
X is: 0.0007130796 -0.0002947929
Iteration: 3
...
...
Value is: -0.9997789
Gradient is: 3.565234 5.995832
X is: 0.0002042478 -8.457939e-05
Value is: -0.9998025
Gradient is: -4.614189 -13.74602
X is: 0.0001822711 -7.553725e-05
Iteration: 4
...
...
Value is: -0.9999917
Gradient is: -382.3041 -921.4625
X is: -6.320081e-06 2.614706e-06
Value is: -0.9999923
Gradient is: -134.0946 -321.2681
X is: -6.921942e-06 2.865841e-06
Iteration: 5
...
...
Value is: -0.9999999
Gradient is: -3446.911 -8320.007
X is: -7.267168e-08 3.009783e-08
Value is: -0.9999999
Gradient is: -3419.361 -8253.501
X is: -7.404627e-08 3.066708e-08
It’s still not perfect, but a lot better.
Finally, let’s go one step further. Can we use torch for constrained optimization?
Quadratic penalty for constrained optimization
In constrained optimization, we still search for a minimum, but that minimum can’t reside just anywhere: Its location has to fulfill some number of additional conditions. In optimization lingo, it has to be feasible .
To illustrate, we stay with the flower function, but add on a constraint: xx has to lie outside a circle of radius sqrt(2), centered at the origin. Formally, this yields the inequality constraint
2−x_1^2−x_2^2<=0
A way to minimize flower and yet, at the same time, honor the constraint is to use a penalty function. With penalty methods, the value to be minimized is a sum of two things: the target function’s output and a penalty reflecting potential constraint violation. Use of a quadratic penalty , for example, results in adding a multiple of the square of the constraint function’s output:
# x^2 + y^2 >= 2
# 2 - x^2 - y^2 <= 0
constraint <- function(x) 2 - torch_square(torch_norm(x))
# quadratic penalty
penalty <- function(x) torch_square(torch_max(constraint(x), other = 0))
A priori, we can’t know how big that multiple has to be to enforce the constraint. Therefore, optimization proceeds iteratively. We start with a small multiplier, 11, say, and increase it for as long as the constraint is still violated:
penalty_method <- function(f, p, x, k_max, rho = 1, gamma = 2, num_iterations = 1) {
for (k in 1:k_max) {
cat("Starting step: ", k, ", rho = ", rho, "\n")
minimize(f, p, x, rho, num_iterations)
cat("Value: ", as.numeric(f(x)), "\n")
cat("X: ", as.matrix(x), "\n")
current_penalty <- as.numeric(p(x))
cat("Penalty: ", current_penalty, "\n")
if (current_penalty == 0) break
rho <- rho * gamma
}
}
minimize() , called from penalty_method() , follows the usual proceedings, but now it minimizes the sum of the target and up-weighted penalty function outputs:
minimize <- function(f, p, x, rho, num_iterations) {
calc_loss <- function() {
optimizer$zero_grad()
value <- f(x) + rho * p(x)
value$backward()
value
}
for (i in 1:num_iterations) {
cat("Iteration: ", i, "\n")
optimizer$step(calc_loss)
}
}
This time, we start from a low-target-loss, but unfeasible value. With yet another change to default L-BFGS (namely, a decrease in tolerance), we see the algorithm exiting successfully after twenty-two iterations, at the point (0.5411692,1.306563) .
x_star <- torch_tensor(c(0.5, 0.5), requires_grad = TRUE)
optimizer <- optim_lbfgs(x_star, line_search_fn = "strong_wolfe", tolerance_change = 1e-20)
penalty_method(flower, penalty, x_star, k_max = 30)
Starting step: 1 , rho = 1
Iteration: 1
Value: 0.3469974
X: 0.5154735 1.244463
Penalty: 0.03444662
Starting step: 2 , rho = 2
Iteration: 1
Value: 0.3818618
X: 0.5288152 1.276674
Penalty: 0.008182613
Starting step: 3 , rho = 4
Iteration: 1
Value: 0.3983252
X: 0.5351116 1.291886
Penalty: 0.001996888
...
...
Starting step: 20 , rho = 524288
Iteration: 1
Value: 0.4142133
X: 0.5411959 1.306563
Penalty: 3.552714e-13
Starting step: 21 , rho = 1048576
Iteration: 1
Value: 0.4142134
X: 0.5411956 1.306563
Penalty: 1.278977e-13
Starting step: 22 , rho = 2097152
Iteration: 1
Value: 0.4142135
X: 0.5411962 1.306563
Penalty: 0
Conclusion
Summing up, we’ve gotten a first impression of the effectiveness of torch ’s L-BFGS optimizer, especially when used with Strong-Wolfe line search. In fact, in numerical optimization – as opposed to deep learning, where computational speed is much more of an issue – there is hardly ever a reason to not use L-BFGS with line search.
We’ve then caught a glimpse of how to do constrained optimization, a task that arises in many real-world applications. In that regard, this post feels a lot more like a beginning than a stock-taking. There is a lot to explore, from general method fit – when is L-BFGS well suited to a problem? – via computational efficacy to applicability to different species of neural networks. Needless to say, if this inspires you to run your own experiments, and/or if you use L-BFGS in your own projects, we’d love to hear your feedback!
Thanks for reading!
Appendix
Rosenbrock function plotting code
library(tidyverse)
a <- 1
b <- 5
rosenbrock <- function(x) {
x1 <- x[1]
x2 <- x[2]
(a - x1)^2 + b * (x2 - x1^2)^2
}
df <- expand_grid(x1 = seq(-2, 2, by = 0.01), x2 = seq(-2, 2, by = 0.01)) %>%
rowwise() %>%
mutate(x3 = rosenbrock(c(x1, x2))) %>%
ungroup()
ggplot(data = df,
aes(x = x1,
y = x2,
z = x3)) +
geom_contour_filled(breaks = as.numeric(torch_logspace(-3, 3, steps = 50)),
show.legend = FALSE) +
theme_minimal() +
scale_fill_viridis_d(direction = -1) +
theme(aspect.ratio = 1)
Flower function plotting code
a <- 1
b <- 1
c <- 4
flower <- function(x) {
a * torch_norm(x) + b * torch_sin(c * torch_atan2(x[2], x[1]))
}
df <- expand_grid(x = seq(-3, 3, by = 0.05), y = seq(-3, 3, by = 0.05)) %>%
rowwise() %>%
mutate(z = flower(torch_tensor(c(x, y))) %>% as.numeric()) %>%
ungroup()
ggplot(data = df,
aes(x = x,
y = y,
z = z)) +
geom_contour_filled(show.legend = FALSE) +
theme_minimal() +
scale_fill_viridis_d(direction = -1) +
theme(aspect.ratio = 1)