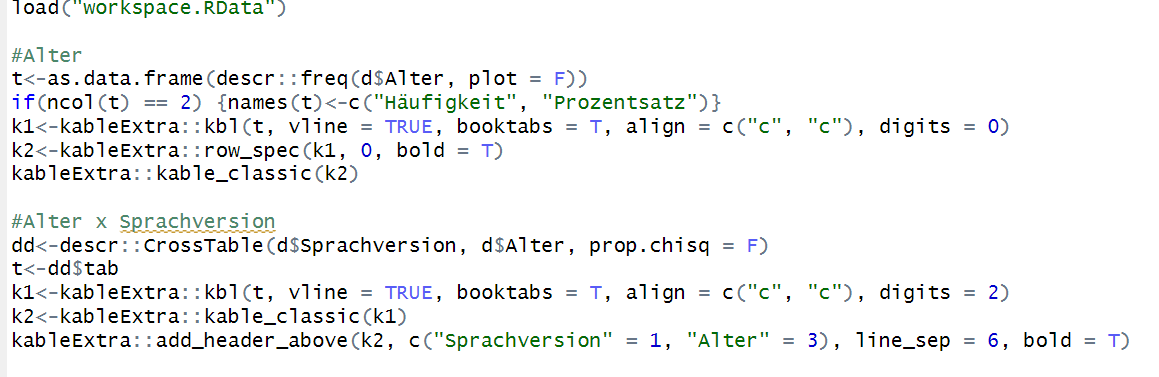
I think I have found the problem. You cannot use vline if you are using booktabs here.
k1<-kableExtra::kbl(t, vline = TRUE, booktabs = T, align = c("c", "c", "c"), digits = 2, row.names = T)
This works
k1 <-kbl(altt , booktabs = T, align = c("c", "c"), digits = 0)
I should have seen it immediately. The author of the {booktabs} package despises vertical lines in a table.
BTW you have a lot of code there that you don't really need. I chopped out some which should make it easier to read. I also renamed some things because it made it easier for me to check things as i worked through the code.
My version of your code. Also availible in Quarto format upon request; I don't remember any Rmarkdown.
library(descr)
library(kableExtra)
# Create data.frame
dat1 <- data.frame(datum = c(1706630620, 1706630638, 1706630657,
1706634604, 1706634668, 1706713347, 1706713462, 1706713517, 1706800923,
1706801182, 1706801418, 1707312101, 1707312342, 1707312639, 1707314100,
1707314172, 1707389396, 1707389732, 1707389801, 1707391257, 1707391620,
1707391988, 1708697243), alter = c(2, 4, 4, 1, 4, 1, 2, 4, 2, 3, 4, 1, 3, 4, 1, 3,
1, 3, 4, 4, 3, 1, 2), sprachversion = c(3, 1, 1, 3, 1, 3,
3, 1, 3, 2, 1, 3, 2, 1, 3, 2, 3, 2, 1, 1, 2, 3, 3) )
#table Alter
altt <- as.data.frame(freq(dat1$alter, plot = FALSE))
names(altt) <- c("Häufigkeit", "Prozentsatz")
k1 <-kbl(altt, vline = TRUE, booktabs = T, align = c("c", "c"), digits = 0)
k2 <-row_spec(k1, 0, bold = T)
kable_classic(k2)
#crosstable Alter x Sprachversion
dd <- CrossTable(dat1$sprachversion, dat1$alter, prop.chisq = F)
t <- dd$tab
k1 <- kableExtra::kbl(t, booktabs = T, align = c("c", "c", "c"), digits = 2, row.names = T)
k2 <- kableExtra::kable_classic(k1)
add_header_above(k2, c("Sprachversion" = 1, "Alter" = 4), line_sep = 6, bold = T)