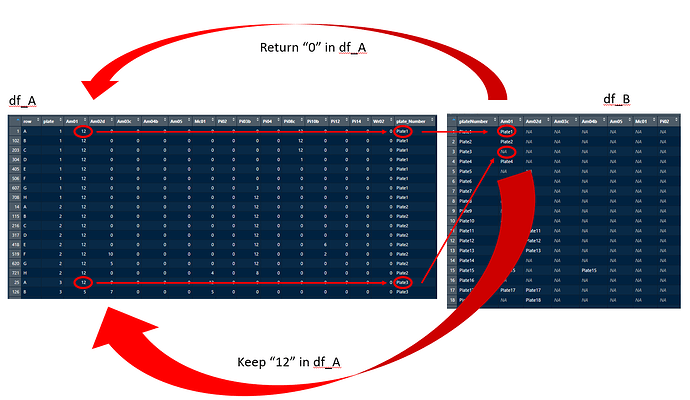
I have two dataframes, df_A and df_B. Below are two smaller examples of the two. The amount of 'Am', 'Pi' and 'Wr' columns is variable per analysis-set. (These are called the 'Markers').
I want the Marker values from df_A replaced by zero if: the plate_Number of column df_A$Plate_Number is present in the respective Marker column of df_B.
If the plate_Number in df_A for a given AM, Pi or Wr column is NA in df_B, the value of df_A should be unchanged.
The columns with the (variable) Am, Pi and Wr values can be referred to in the script using a list.
Markers <- c(Am01, Am02d, Am03c, Am04b, Am05, Mc01, Pi02, Pi03b, Pi04, Pi08c, Pi10b, Pi12, Pi14, Wr02)
Any help would be very appreciated!
>df_A
row plate Am01 Am02d Am03c Am04b Am05 Mc01 Pi02 Pi03b Pi04 Pi08c Pi10b Pi12 Pi14 Wr02 plate_Number
1 A 1 12 0 0 0 0 0 0 0 0 12 0 0 0 0 Plate1
102 B 1 12 0 0 0 0 0 0 0 0 12 0 0 0 0 Plate1
203 C 1 12 0 0 0 0 0 0 0 0 12 0 0 0 0 Plate1
304 D 1 12 0 0 0 0 0 0 0 0 1 0 0 0 0 Plate1
405 E 1 12 0 0 0 0 0 0 0 0 0 0 0 0 0 Plate1
506 F 1 12 0 0 0 0 0 0 0 0 0 0 0 0 0 Plate1
607 G 1 12 0 0 0 0 0 0 3 0 0 0 0 0 0 Plate1
708 H 1 12 0 0 0 0 0 0 12 0 0 0 0 0 0 Plate1
14 A 2 12 0 0 0 0 0 0 12 0 0 0 0 0 0 Plate2
115 B 2 12 0 0 0 0 0 0 12 0 0 0 0 0 0 Plate2
216 C 2 12 0 0 0 0 0 0 12 0 0 0 0 0 0 Plate2
317 D 2 12 0 0 0 0 0 0 12 0 0 0 0 0 0 Plate2
418 E 2 12 0 0 0 0 0 0 12 0 0 6 0 0 0 Plate2
519 F 2 12 10 0 0 0 0 0 12 0 0 2 0 0 0 Plate2
620 G 2 12 5 0 0 0 0 0 12 0 0 0 0 0 0 Plate2
721 H 2 12 0 0 0 0 4 0 8 0 0 0 0 0 0 Plate2
25 A 3 12 0 0 0 0 12 0 0 0 0 0 0 0 0 Plate3
126 B 3 5 7 0 0 0 5 0 0 0 0 0 0 0 0 Plate3
227 C 3 0 12 0 0 0 0 0 0 0 0 0 0 0 0 Plate3
328 D 3 6 6 0 0 0 0 0 0 0 0 0 0 0 0 Plate3
>df_B
plateNumber Am01 Am02d Am03c Am04b Am05 Mc01 Pi02 Pi03b Pi04 Pi08c Pi10b Pi12 Pi14 Wr02
1 Plate1 Plate1 <NA> <NA> <NA> <NA> <NA> <NA> <NA> <NA> <NA> <NA> <NA> <NA> <NA>
2 Plate2 Plate2 <NA> <NA> <NA> <NA> <NA> <NA> Plate2 <NA> <NA> <NA> <NA> <NA> <NA>
3 Plate3 <NA> <NA> <NA> <NA> <NA> <NA> <NA> <NA> <NA> <NA> <NA> <NA> <NA> <NA>
4 Plate4 Plate4 <NA> <NA> <NA> <NA> <NA> <NA> <NA> <NA> <NA> <NA> <NA> <NA> <NA>
5 Plate5 <NA> <NA> <NA> <NA> <NA> <NA> <NA> <NA> <NA> <NA> <NA> <NA> <NA> <NA>
6 Plate6 <NA> <NA> <NA> <NA> <NA> <NA> <NA> <NA> <NA> <NA> <NA> Plate6 <NA> <NA>
7 Plate7 <NA> <NA> <NA> <NA> <NA> <NA> <NA> <NA> <NA> <NA> Plate7 <NA> <NA> <NA>
8 Plate8 <NA> <NA> <NA> <NA> <NA> <NA> <NA> <NA> <NA> <NA> Plate8 <NA> <NA> <NA>
9 Plate9 <NA> <NA> <NA> <NA> <NA> <NA> <NA> <NA> <NA> <NA> <NA> <NA> Plate9 <NA>
10 Plate10 <NA> <NA> <NA> <NA> <NA> <NA> <NA> <NA> <NA> <NA> <NA> <NA> <NA> <NA>
11 Plate11 Plate11 Plate11 <NA> <NA> <NA> <NA> <NA> <NA> <NA> <NA> <NA> <NA> <NA> <NA>
12 Plate12 Plate12 Plate12 <NA> <NA> <NA> <NA> <NA> <NA> <NA> <NA> <NA> <NA> <NA> <NA>
13 Plate13 Plate13 Plate13 <NA> <NA> <NA> <NA> <NA> <NA> <NA> <NA> <NA> <NA> <NA> <NA>
14 Plate14 Plate14 <NA> <NA> <NA> <NA> <NA> <NA> <NA> <NA> <NA> <NA> <NA> <NA> <NA>
15 Plate15 Plate15 <NA> <NA> Plate15 <NA> <NA> <NA> <NA> <NA> <NA> <NA> <NA> <NA> <NA>
16 Plate16 <NA> <NA> <NA> <NA> <NA> <NA> <NA> <NA> <NA> <NA> <NA> <NA> <NA> <NA>
17 Plate17 Plate17 Plate17 <NA> <NA> <NA> <NA> <NA> <NA> <NA> <NA> <NA> <NA> <NA> <NA>
18 Plate18 <NA> Plate18 <NA> <NA> <NA> <NA> <NA> <NA> <NA> <NA> <NA> <NA> <NA> <NA>
19 Plate19 <NA> Plate19 <NA> <NA> <NA> <NA> <NA> <NA> <NA> <NA> <NA> <NA> <NA> <NA>
20 Plate20 <NA> Plate20 <NA> <NA> <NA> <NA> <NA> <NA> <NA> <NA> <NA> <NA> <NA> <NA>