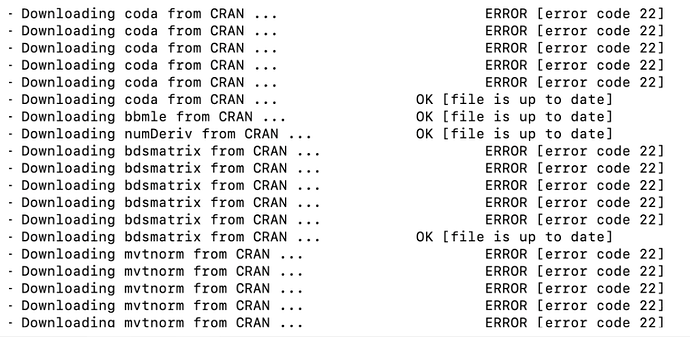
I am using renv to manage R packages in a project. The project uses packages both from bioconductor and CRAN. When I update/install packages from a lock file with "renv::restore()", I receive the following error message:
For certain CRAN packages, it attempts multiple downloads before finally succeeding. Running with "options(renv.download.trace = TRUE); renv::restore()" gives the following, more detailed output:
- Downloading pillar from CRAN ...
# Downloading 'https://bioconductor.org/packages/3.14/bioc/src/contrib/Archive/pillar/pillar_1.9.0.tar.gz' [curl] --------
## Request -------------------------------------------------------------------
location
fail
silent
show-error
user-agent = "renv (1.0.11); R (4.1.3 x86_64-apple-darwin13.4.0 x86_64 darwin13.4.0)"
url = "https://bioconductor.org/packages/3.14/bioc/src/contrib/Archive/pillar/pillar_1.9.0.tar.gz"
output = "/Users/thomaswalter/Library/Caches/org.R-project.R/R/renv/source/repository/pillar/renv-tempfile-15df37e7458c"
connect-timeout = "20"
retry = "3"
## Output --------------------------------------------------------------------
curl: (22) The requested URL returned error: 404
## Status --------------------------------------------------------------------
22
ERROR [error code 22]
- Downloading pillar from CRAN ...
# Downloading 'https://bioconductor.org/packages/3.14/data/annotation/src/contrib/Archive/pillar/pillar_1.9.0.tar.gz' [curl] --------
## Request -------------------------------------------------------------------
location
fail
silent
show-error
user-agent = "renv (1.0.11); R (4.1.3 x86_64-apple-darwin13.4.0 x86_64 darwin13.4.0)"
url = "https://bioconductor.org/packages/3.14/data/annotation/src/contrib/Archive/pillar/pillar_1.9.0.tar.gz"
output = "/Users/thomaswalter/Library/Caches/org.R-project.R/R/renv/source/repository/pillar/renv-tempfile-15df352d0cf26"
connect-timeout = "20"
retry = "3"
## Output --------------------------------------------------------------------
curl: (22) The requested URL returned error: 404
## Status --------------------------------------------------------------------
22
ERROR [error code 22]
- Downloading pillar from CRAN ...
# Downloading 'https://bioconductor.org/packages/3.14/data/experiment/src/contrib/Archive/pillar/pillar_1.9.0.tar.gz' [curl] --------
## Request -------------------------------------------------------------------
location
fail
silent
show-error
user-agent = "renv (1.0.11); R (4.1.3 x86_64-apple-darwin13.4.0 x86_64 darwin13.4.0)"
url = "https://bioconductor.org/packages/3.14/data/experiment/src/contrib/Archive/pillar/pillar_1.9.0.tar.gz"
output = "/Users/thomaswalter/Library/Caches/org.R-project.R/R/renv/source/repository/pillar/renv-tempfile-15df310ea90f0"
connect-timeout = "20"
retry = "3"
## Output --------------------------------------------------------------------
curl: (22) The requested URL returned error: 404
## Status --------------------------------------------------------------------
22
ERROR [error code 22]
- Downloading pillar from CRAN ...
# Downloading 'https://bioconductor.org/packages/3.14/workflows/src/contrib/Archive/pillar/pillar_1.9.0.tar.gz' [curl] --------
## Request -------------------------------------------------------------------
location
fail
silent
show-error
user-agent = "renv (1.0.11); R (4.1.3 x86_64-apple-darwin13.4.0 x86_64 darwin13.4.0)"
url = "https://bioconductor.org/packages/3.14/workflows/src/contrib/Archive/pillar/pillar_1.9.0.tar.gz"
output = "/Users/thomaswalter/Library/Caches/org.R-project.R/R/renv/source/repository/pillar/renv-tempfile-15df3188e4e57"
connect-timeout = "20"
retry = "3"
## Output --------------------------------------------------------------------
curl: (22) The requested URL returned error: 404
## Status --------------------------------------------------------------------
22
ERROR [error code 22]
- Downloading pillar from CRAN ...
# Downloading 'https://bioconductor.org/packages/3.14/books/src/contrib/Archive/pillar/pillar_1.9.0.tar.gz' [curl] --------
## Request -------------------------------------------------------------------
location
fail
silent
show-error
user-agent = "renv (1.0.11); R (4.1.3 x86_64-apple-darwin13.4.0 x86_64 darwin13.4.0)"
url = "https://bioconductor.org/packages/3.14/books/src/contrib/Archive/pillar/pillar_1.9.0.tar.gz"
output = "/Users/thomaswalter/Library/Caches/org.R-project.R/R/renv/source/repository/pillar/renv-tempfile-15df32001d31d"
connect-timeout = "20"
retry = "3"
## Output --------------------------------------------------------------------
curl: (22) The requested URL returned error: 404
## Status --------------------------------------------------------------------
22
ERROR [error code 22]
- Downloading pillar from CRAN ...
# Downloading 'https://lib.stat.cmu.edu/R/CRAN/src/contrib/Archive/pillar/pillar_1.9.0.tar.gz' [curl] --------
## Request -------------------------------------------------------------------
location
fail
silent
show-error
user-agent = "renv (1.0.11); R (4.1.3 x86_64-apple-darwin13.4.0 x86_64 darwin13.4.0)"
url = "https://lib.stat.cmu.edu/R/CRAN/src/contrib/Archive/pillar/pillar_1.9.0.tar.gz"
output = "/Users/thomaswalter/Library/Caches/org.R-project.R/R/renv/source/repository/pillar/renv-tempfile-15df33d4dbbbf"
connect-timeout = "20"
retry = "3"
## Output --------------------------------------------------------------------
[no output generated]
## Status --------------------------------------------------------------------
0
OK [434.1 Kb in 0.29s]
It seems it tries to locate the package at four different bioconductor links before finally attempting CRAN. This only happens with CRAN packages, and does not happen with all of the CRAN packages. How can I configure renv to always search CRAN packages in CRAN on first attempt? Thanks in advance and let me know any other info I can give