Hello,
I am inserting a reprex of the minimal app I can get to demonstrate the problem and a reprex of the script called by the app, together with a reprext of the program that creates the files. The problem I am having is that the statement "names_files <- list.files(getwd(), pattern = "[0-9].csv", all.files = FALSE, full.names = TRUE, recursive = FALSE, ignore.case = FALSE, include.dirs = FALSE, no.. = TRUE)" at the end of the script called by the app returns the error "Error: $ operator is invalid for atomic vectors". However if i write that statement at the beginning of the script or inside the outermost loop, the error does not occur. Could you please run these programs and see if the same thing happens in your environment. I know this is a different question, but I cannot test the file.rename(function) is I cannot list the directory.
Thanks,
APP
library(shiny)
library(shinyjs)
#>
#> Attaching package: 'shinyjs'
#> The following object is masked from 'package:shiny':
#>
#> runExample
#> The following objects are masked from 'package:methods':
#>
#> removeClass, show
ui <- fluidPage(
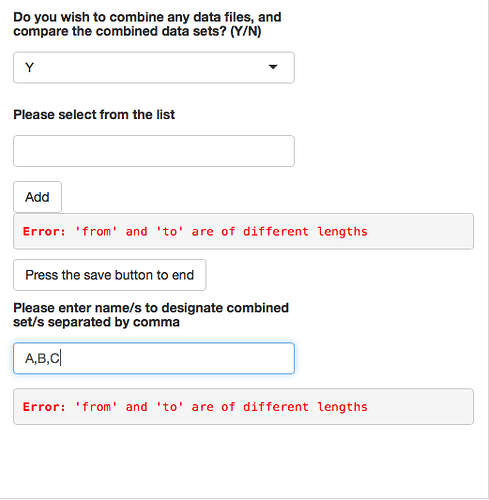
selectInput("combinefiles", label = h5(strong("Do you wish to combine any data files, and compare the combined data sets? (Y/N) ")),
choices = c("", "Y", "N"),selected = NULL),
verbatimTextOutput("combiningFiles"),
verbatimTextOutput("combineChosen"),
#verbatimTextOutput("filesToCombine"),
useShinyjs(),
conditionalPanel(
condition = "output.toCombine > '0'",
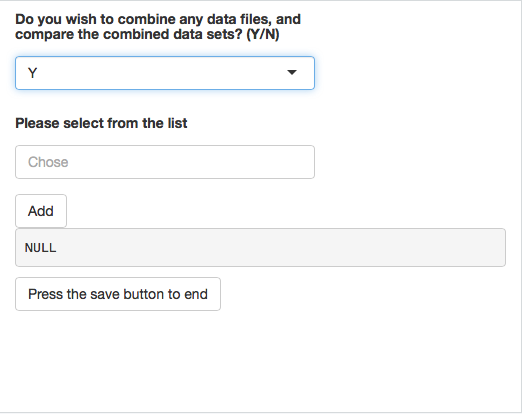
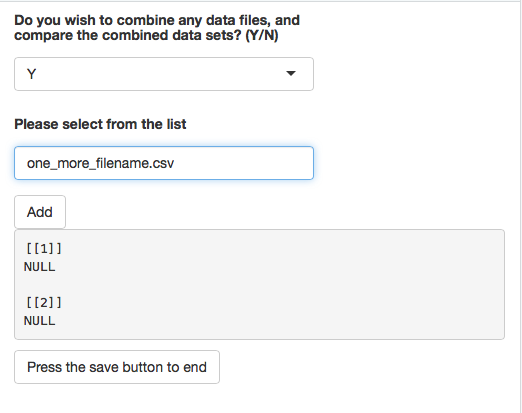
selectInput(inputId = "select",label = h5(strong("Please select from the list")), choices = c(Chose = "", list.files("~/Development/glycoPipeApp")), multiple = TRUE, selectize = TRUE)
),
conditionalPanel(
condition = "output.displayAddButton > '0'",
actionButton('add','Add')
),
verbatimTextOutput("samelist"),
conditionalPanel(
condition = "output.displayAddButton == 1",
actionButton("sBtn", "Press the save button to end")
),
conditionalPanel(
condition = "output.displayTheSaveButton > '0'",
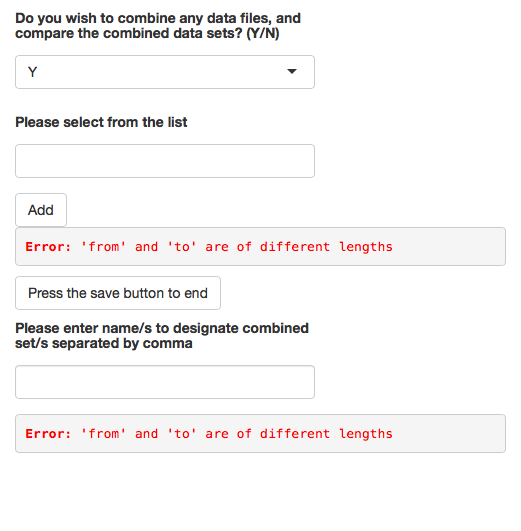
textInput("textbox", h5(strong("Please enter name/s to designate combined set/s separated by comma")))),
strong(verbatimTextOutput("list")),
verbatimTextOutput("caption")
)
#selections = NULL,
glycoPipe <- function(response = NULL, fOfData = NULL, combineResult = NULL, listContents = NULL, vals = NULL){
enteredValue = NULL
nameList = NULL
answer = NULL
fileChoice = NULL
combination = NULL
combinations = NULL
comb = NULL
nameListSize = NULL
choseDataFiles = NULL
if(!is.null(response)){
answer = response
}
if(!is.null(fOfData)){
fileChoice = fOfData
}
if(!is.null(combineResult)){
combination = combineResult
}
if(!is.null(listContents)){
enteredValue = listContents
}
if(!is.null(vals)){
nameList <- vals
}
glyCount1(answer, fileChoice, combination, enteredValue, nameList)
}
server <- function(input, output, session){
listContents = NULL
source("Utilities/utilities.R")
source("glycoPipeFunctions/glycoPipe_fcns.R")
source("glyCount1.R")
output$toCombine <- reactive({
req(input$combinefiles)
return(length(input$combinefiles))
})
outputOptions(output, "toCombine", suspendWhenHidden = FALSE)
output$displayAddButton <- reactive({
req(input$combinefiles)
return(length(input$combinefiles))
})
outputOptions(output, "displayAddButton", suspendWhenHidden = FALSE)
output$displayTheSaveButton <- reactive({
req(input$sBtn)
return(input$sBtn)
})
outputOptions(output, "displayTheSaveButton", suspendWhenHidden = FALSE)
myValues <- reactiveValues()
observe({
if(input$add > 0){
myValues$dList <- c(isolate(myValues$dList), isolate(list(input$select)))
}
})
# #unlist(input$filescombine)
output$samelist<-renderPrint({
#listContents <- list()
listContents <- append(listContents, myValues$dList)
print(listContents)
if(input$sBtn > 0){
numberOfSelectedSets <- glycoPipe(response = NULL, fOfData = NULL, combineResult = NULL , listContents)
paste("Please enter", numberOfSelectedSets$nameListSize, "names to designate your", numberOfSelectedSets$nameListSize, "sets")
}
})
VALUES <- list()
observe({
isolate({
req(input$textbox)
VALUES <- input$textbox
VALUES <- append(VALUES, list(input$textbox))
updateTextInput(session, inputId = "textbox", value = VALUES)
})
})
output$caption <- renderPrint({
vals <- list()
vals <- append(vals, input$textbox)
if(input$sBtn > 0){
result <- glycoPipe(response = NULL, fOfData = NULL, combineResult = NULL, listContents = NULL, vals)
#unlist(input$filescombine)
}
})
session$allowReconnect(TRUE)
}
shinyApp(ui = ui, server = server)
SCRIPT CALLED BY THE APP
ibrary(stringi)
glyCount1 <- function(answer = NULL, fileChoice = NULL, combination = NULL, enteredValue = NULL, nameList) {
# Length of list
lc <- enteredValue
choseDataFiles = TRUE
fileLength <- length(lc)
first_path <- NULL
new_path <- NULL
old_path <- NULL
file_content <- NULL
names_files <- NULL
for(i in 1:length(lc)){
for(j in 1:length(lc[[i]])){
if(!is.null(lc[[i]])){
first_path[[j]]<- paste(getwd(), "/", lc[[i]][j], sep = "")
file_content[[j]] <- read.csv(file = first_path[[i]], header = TRUE, sep = ",")
old_path[[j]] <- paste(getwd(), "/", i, ".csv", sep = "")
write.table(file_content[[j]], file = old_path[[j]], append = TRUE, col.names = FALSE)
}
}
new_dataFns <- as.list(unlist(strsplit(as.character(nameList), ",")))
for(i in 1:length(new_dataFns)){
new_path[i] <- gsub(" ", "", paste(getwd(), "/", new_dataFns[i], sep = ""))
}
}
names_files <- list.files(getwd(), pattern = "[0-9].csv", all.files = FALSE, full.names = TRUE, recursive = FALSE, ignore.case = FALSE, include.dirs = FALSE, no.. = TRUE)
#apply(1:length(new_path), function(i){ file.rename(names_files[[i]], new_path[[i]]) })
#file.rename(names_files, new_path)
}
Created on 2018-09-19 by the reprex package (v0.2.1)
SCRIPT TO CREATE FILES
library(magrittr)
library(reprex)
JF_160426_Dep2Plas_tryp_Gpep_SIDtargPSM<- NULL
JF_160426_Dep2Plas_tryp_Gpep_SIDtargPSM %>% # table that should become a file
head(5) %>% # 5 lines might be plenty?
dput()
#> NULL
0
#> [1] 0
JF_160426_Dep2Plas_tryp_Gpep_SIDtargPSM <-
# Everything below here is the output from above. I used
# fake data for this.
structure(list(
confidence = c("High", "High", "High", "High", "High", "High", "Medium", "High", "High", "High", "High", "High", "High", "High", "High", "Medium", "High", "High", "Medium","High"
),
deltaScore = c("0.1667", "0.5789", "1", "0.56", "0.52", "1", "0.56", "0.5882", "1", "0.52", "1", "0.56", "0.5", "0.2857", "0.5556", "0.55", "0", "0.2857", "0.5789", "0.5"
),
PSM_ambiguity = c("Unambiguous", "Unambiguous", "Unambiguous", "Unambiguous", "Unambiguous", "Unambiguous", "Unambiguous", "Unambiguous", "Unambiguous", "Unambiguous", "Unambiguous", "Unambiguous", "Unambiguous", "Unambiguous", "Unambiguous", "Unambiguous", "Ambiguous", "Unambiguous", "Unambiguous", "Medium"
),
Annotated_Sequence = c("InHcR", "InHcR", "InHcR", "InHcR", "InHcR", "InHcR", "InHcR", "EnGTVSR", "InHcR", "InHcR", "InHcR", "InHcR", "InHcR", "InHcR", "InHcR", "InHcR", "InHcR", "InHcR", "InHcR", "InHcR"
),
Modifications = c("N2(HexNAc); C4(Carbamidomethyl)", "N2(HexNAc); C4(Carbamidomethyl)", "N2(HexNAc); C4(Carbamidomethyl)", "N2(HexNAc); C4(Carbamidomethyl)", "N2(HexNAc); C4(Carbamidomethyl)","N2(HexNAc) C4(Carbamidomethyl)", "N2(HexNAc); C4(Carbamidomethyl)", "N2(HexNAc); C4(Carbamidomethyl)", "N2(HexNAc); C4(Carbamidomethyl)", "N2(HexNAc); C4(Carbamidomethyl)", "N2(HexNAc); C4(Carbamidomethyl)", "N2(HexNAc); C4(Carbamidomethyl)", "N2(HexNAc); C4(Carbamidomethyl)", "N2(HexNAc); C4(Carbamidomethyl)", "N2(HexNAc); C4(Carbamidomethyl)", "N2(HexNAc); C4(Carbamidomethyl)", "N2(HexNAc); C4(Carbamidomethyl)","N2(HexNAc); C4(Carbamidomethyl", "N2(HexNAc); C4(Carbamidomethyl", "N2(HexNAc); C4(Carbamidomethyl)"
)
), .Names = c("confidence", "deltaScore","PMS_Ambiguity", "Annotated_Sequence", "Modifications"), row.names = c(
NA,
-20L
), class = c("tbl_df", "tbl", "data.frame"))
write.csv(JF_160426_Dep2Plas_tryp_Gpep_SIDtargPSM, "F_160426_Dep2Plas_tryp_Gpep_SIDtargPSM.csv")
rm(JF_160426_Dep2Plas_tryp_Gpep_SIDtargPSM)
Created on 2018-09-19 by the reprex package (v0.2.1)