Hello,
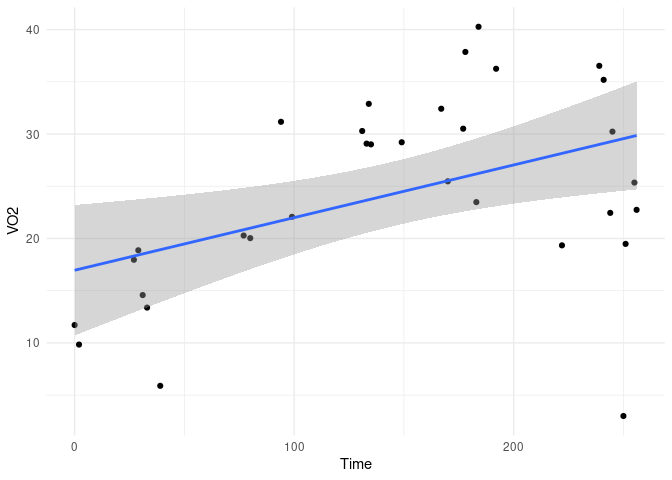
I have a large data set that analyzes exercising subjects' oxygen consumption over time (x= Time, y = VO2). This data fits a monoexponential function.
Here is a brief, sample data frame:
'''
VO2 <- c(11.71,9.84,17.96,18.87,14.58,13.38,5.89,20.28,20.03,31.17,22.07,30.29,29.08,32.89,29.01,29.21,32.42,25.47,30.51,37.86,23.48,40.27,36.25,19.34,36.53,35.19,22.45,30.23,3,19.48,25.35,22.74)
Time <- c(0,2,27,29,31,33,39,77,80,94,99,131,133,134,135,149,167,170,177,178,183,184,192,222,239,241,244,245,250,251,255,256)
DF <- data.frame(VO2,Time)
'''
My primary question is the following:
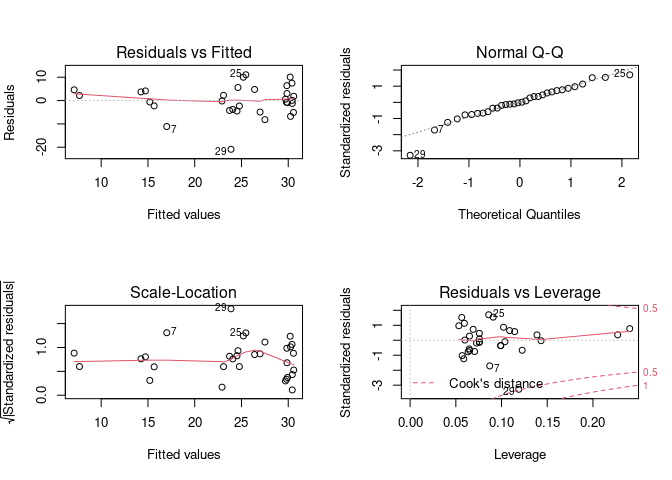
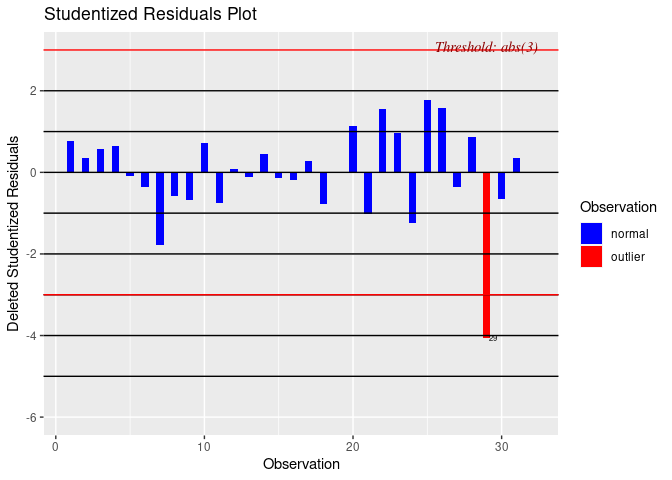
This data can be messy at points and needs to be cleaned. Using R, how do I remove outliers that occur +/- 3 standard deviations from the mean of a monoexponential function? Each data set I clean will be slightly different, so I would like R to fit some monoexponential function to the data and remove (replace with NA) all the outlying values. I then can continue manipulating the data to achieve my goal.
So far, I have this (unsuccessful) code to fit a function to the above data. Not only does it not fit well, but I am also unable to create a smooth function.
'''
fit <- lm(VO2~poly(Time,2,raw=TRUE))
xx <- seq(1,250, length=32)
plot(Time,VO2,pch=19,ylim=c(0,50))+
lines(xx, predict(fit, data.frame(DF=xx)), col="red")
'''
Thank you for the comments and valuable feedback. As I continue to learn and research, I will add to this post with successful/less successful attempts at the code for this process. Thank you for your knowledge, assistance and understanding.