Hello! My situation is that I am able to run a set of code in R and produce plots using ggplot2 without specifying dropping N/A values. Its doing it in the background somehow. I am working on putting everything into a markdown file and at this particular set of code it isnt removing the n/a values for the data frame and producing the plots without n/a. In r markdown Im able to get plots but now it gives me a section in the bar chart with n/a. Any ideaswhy this is happening and how I can get r markdown to produce what I am getting in R?
This is the original dataframe with output below
dput(head(dailymerged, 5))
structure(list(Id = c(1503960366, 1503960366, 1503960366, 1503960366,
1503960366), Date = structure(c(16903, 16904, 16906, 16907, 16908
), class = "Date"), Weekday = structure(c(3L, 4L, 6L, 7L, 1L), .Label = c("Sun",
"Mon", "Tue", "Wed", "Thu", "Fri", "Sat"), class = c("ordered",
"factor")), Calories = c(1985L, 1797L, 1745L, 1863L, 1728L),
TotalSteps = c(13162L, 10735L, 9762L, 12669L, 9705L), TotalDistance = c(8.5,
6.96999979019165, 6.28000020980835, 8.15999984741211, 6.48000001907349
), VeryActiveDistance = c(1.87999999523163, 1.57000005245209,
2.14000010490417, 2.71000003814697, 3.19000005722046), ModeratelyActiveDistance = c(0.550000011920929,
0.689999997615814, 1.25999999046326, 0.409999996423721, 0.779999971389771
), LightlyActiveDistance = c(6.05999994277954, 4.71000003814697,
2.82999992370605, 5.03999996185303, 2.50999999046326), SedentaryDistance = c(0,
0, 0, 0, 0), VeryActiveMinutes = c(25L, 21L, 29L, 36L, 38L
), ModeratelyActiveMinutes = c(13L, 19L, 34L, 10L, 20L),
LightlyActiveMinutes = c(328L, 217L, 209L, 221L, 164L), SedentaryMinutes = c(728L,
776L, 726L, 773L, 539L), TotalSleepRecords = c(1L, 2L, 1L,
2L, 1L), TotalMinutesAsleep = c(327L, 384L, 412L, 340L, 700L
), TotalTimeInBed = c(346L, 407L, 442L, 367L, 712L)), row.names = c(NA,
5L), class = "data.frame")
From dailymerged I created another dataframe named ActivityLong.
structure(list(Id = c(1503960366, 1503960366, 1503960366, 1503960366,
1503960366), Date = structure(c(16903, 16903, 16903, 16903, 16904
), class = "Date"), Weekday = structure(c(3L, 3L, 3L, 3L, 4L), .Label = c("Sun",
"Mon", "Tue", "Wed", "Thu", "Fri", "Sat"), class = c("ordered",
"factor")), Calories = c(1985L, 1985L, 1985L, 1985L, 1797L),
TotalSteps = c(13162L, 13162L, 13162L, 13162L, 10735L), TotalDistance = c(8.5,
8.5, 8.5, 8.5, 6.96999979019165), Level = structure(c(4L,
3L, 2L, 1L, 4L), .Label = c("Sedentary", "LightlyActive",
"ModeratelyActive", "VeryActive"), class = "factor"), Distance = c(1.87999999523163,
0.550000011920929, 6.05999994277954, 0, 1.57000005245209),
Minutes = c(25, 13, 328, 728, 21), TotalSleepRecords = c(1L,
1L, 1L, 1L, 2L), TotalMinutesAsleep = c(327L, 327L, 327L,
327L, 384L), TotalTimeInBed = c(346L, 346L, 346L, 346L, 407L
)), row.names = c(NA, -5L), class = c("tbl_df", "tbl", "data.frame"
))
Here is the code I run to get ActivityLong
dailymerged <- dailymerged |>
rename(LightlyActiveDistance=LightActiveDistance,
ModeratelyActiveMinutes=FairlyActiveMinutes,
SedentaryDistance=SedentaryActiveDistance)
ActivityLong <- dailymerged |>
pivot_longer(VeryActiveDistance:SedentaryMinutes, names_to = "Level_Metric")
head(ActivityLong, 3)
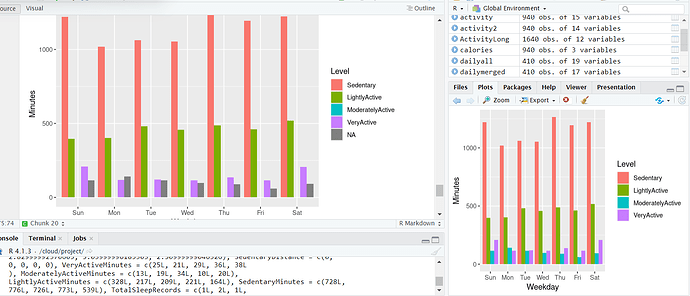
Running this in R ends up giving me ActivityLong with 1640 obs of 12 variables and then seems to be using that to make the plots below. But, in R markdown I cant get the dataframe to automatically drop the n/a values and its giving me 2-3000 obs and then making the plots which produce a warning of removing X # of row with n/a values but then the pplots have the n/a values in them. See below.
This is the code I am running to get plots in R.
ggplot(ActivityLong, aes(x= Weekday, y= Minutes, fill=Level)) +
geom_col()
ggplot(ActivityLong, aes(x = Weekday, y = Minutes, fill=Level)) +
geom_col(position="dodge")
The plot in the middle is what I am getting in R markdown and the plot to the R is what I am getting in R. How do I get R markdown to produce the same plot as in R?
Thanks.