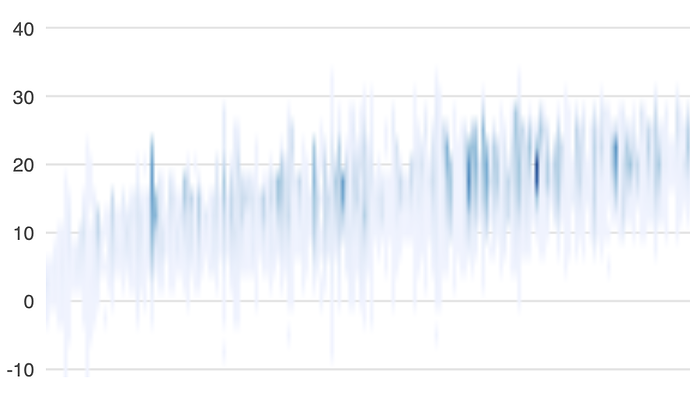
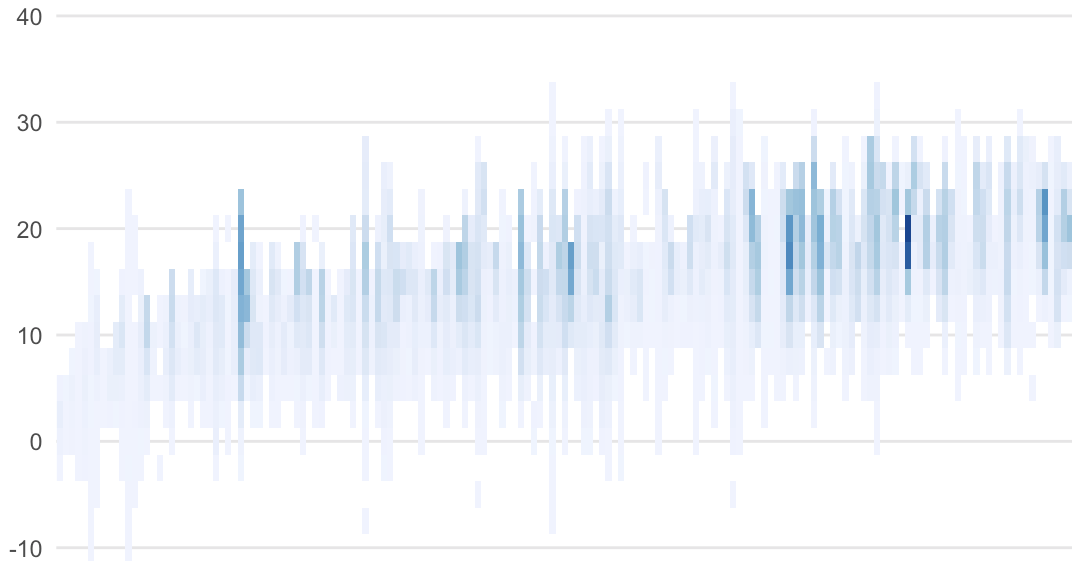
I've a plot in a .qmd document. The plot has many (raster) segments that appear very soft and blurry when converting the document to PDF.
Here's a screenshot of the rendered graph:
And how it looks in R Studio:
How can avoid this so that the rendered figure looks the same as in R?
I'm using Quarto 1.5.56 on macOS 13.6.7.
Below is a reproducible example with a subset of my data.
---
format: pdf
execute:
echo: false
warning: false
knitr:
opts_chunk:
dev: cairo_pdf
---
```{r}
#| include: false
df <- structure(list(x = c("Alps", "Alps", "Alps", "Alps", "Alps",
"Alps", "Alps", "Alps", "Caucasus Mountains", "Cascade Range",
"Alps", "Himalayas", "Tian Shan", "Himalayas", "Caucasus Mountains",
"Borneo", "Himalayas", "Cordillera Oriental (Northern Andes)",
"Alps", "Tian Shan", "Caucasus Mountains", "Albertine Rift",
"Tian Shan", "Tian Shan", "Central American Cordillera", "Southwest Basins and Ranges",
"Southwest Basins and Ranges", "Cascade Range", "Transverse Ranges",
"Transverse Ranges", "Southwest Basins and Ranges", "Transverse Ranges",
"Trans-Mexican Volcanic Belt", "Trans-Mexican Volcanic Belt",
"Trans-Mexican Volcanic Belt", "Great Basin Ranges", "Hawaiian Islands",
"Southwest Basins and Ranges", "Transverse Ranges", "Transverse Ranges",
"Dry Andes", "Southwest Basins and Ranges", "Great Basin Ranges",
"Cascade Range", "Cordillera Occidental (Northern Andes)", "Great Basin Ranges",
"Transverse Ranges", "Southwest Basins and Ranges", "Sierra Madre del Sur",
"Central American Cordillera", "Hawaiian Islands", "Cordillera Oriental (Northern Andes)",
"Cordillera Oriental (Central Andes)", "Cascade Range", "Great Basin Ranges",
"Cascade Range", "Trans-Mexican Volcanic Belt", "Central American Cordillera",
"Sierra Madre del Sur", "Cascade Range", "Sierra Madre del Sur",
"Caucasus Mountains", "Sierra Madre del Sur", "Cascade Range",
"Southwest Basins and Ranges", "Hawaiian Islands", "Cascade Range",
"Cordillera Oriental (Northern Andes)", "Central American Cordillera",
"Hawaiian Islands", "Southwest Basins and Ranges", "Cordillera Oriental (Northern Andes)",
"Sierra Madre del Sur", "Hawaiian Islands", "Cordillera Oriental (Northern Andes)",
"Great Basin Ranges", "Hawaiian Islands", "Southwest Basins and Ranges",
"Hawaiian Islands", "Trans-Mexican Volcanic Belt", "Cordillera Occidental (Central Andes)",
"Cordillera Occidental (Northern Andes)", "Cordillera Central (Northern Andes)",
"Southwest Basins and Ranges", "Great Basin Ranges", "Transverse Ranges",
"Cordillera Oriental (Northern Andes)", "Cordillera Occidental (Northern Andes)",
"Great Basin Ranges", "Trans-Mexican Volcanic Belt", "Transverse Ranges",
"Dry Andes", "Great Basin Ranges", "Cordillera Occidental (Northern Andes)",
"Caucasus Mountains", "Central American Cordillera", "Cordillera Occidental (Central Andes)",
"Caucasus Mountains", "Caucasus Mountains", "Gregory Rift"),
y = c(7.5, 15, 17.5, 5, 12.5, 10, 22.5, 20, 20,
15, 2.5, 22.5, 22.5, 20, 22.5, 25, 27.5, 12.5, 0, 12.5, 15,
20, 15, 7.5, 25, 30, 27.5, 12.5, 25, 22.5, 20, 20, 15, 22.5,
17.5, 30, 15, 32.5, 17.5, 15, 12.5, 15, 32.5, 17.5, 12.5,
25, 30, 25, 25, 15, 17.5, 25, 7.5, 5, 22.5, 10, 20, 20, 22.5,
27.5, 27.5, 17.5, 17.5, 20, 22.5, 25, 7.5, 20, 22.5, 22.5,
17.5, 15, 20, 7.5, 7.5, 27.5, 12.5, 35, 20, 10, 25, 5, 17.5,
12.5, 15, 12.5, 22.5, 15, 20, 12.5, 27.5, 15, 35, 10, 10,
27.5, 5, 7.5, 12.5, 22.5), n = c(2114L, 3673L, 4012L, 1223L,
3211L, 2799L, 2512L, 3751L, 1942L, 1842L, 353L, 277L, 535L,
438L, 1786L, 429L, 312L, 1846L, 49L, 365L, 1352L, 439L, 480L,
260L, 2905L, 1405L, 1524L, 1101L, 853L, 1663L, 1330L, 1693L,
1734L, 1722L, 2207L, 671L, 124L, 736L, 1291L, 889L, 470L,
476L, 448L, 2086L, 1081L, 896L, 312L, 1758L, 1239L, 1409L,
368L, 644L, 363L, 245L, 1181L, 773L, 2380L, 2422L, 1057L,
312L, 946L, 1508L, 1319L, 1081L, 1756L, 724L, 543L, 904L,
2479L, 914L, 890L, 1132L, 1402L, 37L, 473L, 717L, 110L, 185L,
471L, 263L, 110L, 158L, 1010L, 149L, 810L, 193L, 1084L, 908L,
1180L, 813L, 413L, 564L, 225L, 635L, 944L, 1082L, 281L, 457L,
1206L, 244L)), class = c("tbl_df", "tbl", "data.frame"), row.names = c(NA,
-100L))
library(ggplot2)
```
```{r}
df |>
dplyr::mutate(x = forcats::fct_reorder(factor(x), .x = y)) |>
ggplot(aes(x = x, y = y, fill = n)) +
geom_raster() +
scale_fill_distiller(
palette = "Blues",
trans = "reverse",
guide = guide_colorbar(reverse = TRUE)
) +
theme_classic() +
theme(
panel.grid.major.y = element_line(color = "grey90"),
axis.line = element_blank(),
axis.ticks = element_blank(),
axis.text.x = element_blank()
)
```