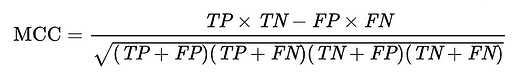Dear R Studio Community,
I am running a multilevel binary logistic regression (MLBLR) model using glmer. After having trained the MLBLR on the training data (which was created using Tidymodels), I now intend to tune/calibrate the prediction probability threshold. I hereby want to maximise Matthews correlation coefficient (MCC). Do you have an idea how I could best do this when building upon the following code?
I tried it via a for loop in which I tried recreating the MCC formula for each cutoff value. However, when plotting the results for the different cutoffs I cannot distinguish a fitting cutoff value, even when I have more detailed data.
MCC formula:
MCC = (TPTN-FPFN)/sqrt((TP+FP)(TP+FN)(TN+FP)(TN+FN))
Code:
(Reproducable Example):
Df <- tibble::tribble(
~year_week,~Runner,~NewRRI,~Distance, ~HR, ~Gender,~Age,~BMI,~PreviousRRI,
"2019-41", "M01" , 0, 5000, 120, "Male", 23, 18, 1,
"2019-41", "M02" , 0, 6000, 125,"Female", 36, 20, 0,
"2019-41", "M03" , 0, 8000, 130, "Male", 56, 21, 0,
"2019-42", "M01" , 0, 5500, 122, "Male", 23, 18, 1,
"2019-42", "M02" , 0, 7000, 128,"Female", 36, 20, 0,
"2019-42", "M03" , 0, 15000, 132, "Male", 56, 21, 0,
"2019-43", "M01" , 1, 3000, 120, "Male", 23, 18, 1,
"2019-43", "M02" , 0, 9000, 127,"Female", 36, 20, 0,
"2019-43", "M03" , 0, 9500, 131, "Male", 56, 21, 0,
"2019-44", "M01" , 0, 15000, 125, "Male", 23, 18, 1,
"2019-44", "M02" , 0, 9000, 127,"Female", 36, 20, 0,
"2019-44", "M03" , 0, 9500, 131, "Male", 56, 21, 0,
) %>%
mutate(Gender = as.factor(Gender),
PreviousRRI = as.factor(PreviousRRI),
NewRRI = as.factor(NewRRI),
Runner = as.factor(Runner))
library(tidyverse)
library(tidymodels)
library(multilevelmod)
library(themis)
library(caret)
library(lme4)
# Split data
Df <- Df %>% arrange(year_week)
Df_splits <- initial_time_split(Df, prop = 0.8)
RunningData_train <- training(Df_splits)
RunningData_test <- testing(Df_splits)
# Rescale
RunningData_train_norm <- preProcess(RunningData_train)
RunningData_train <- predict(RunningData_train_norm, RunningData_train)
RunningData_test <- predict(RunningData_train_norm, RunningData_test)
# MLBLR model
MLBR_L <- glmer(NewRRI ~ Distance + HR + Gender + Age + BMI + PreviousRRI + (1|Runner),
data = RunningData_train,
family = binomial(link = "logit"),
control = glmerControl(optimizer = "bobyqa"),
nAGQ = 1)
# Tune probability value
cutoffs <- seq(0.1,0.9,0.1)
mcc <- NULL
for (i in seq(along = cutoffs)){
prediction <- ifelse(MLBR_L@resp[[".->mu"]] >=cutoffs[i],1,0) # Predictions
mcc <- c(mcc, (length(which(RunningData_train$NewRRI == prediction) && which(RunningData_train$NewRRI == 1))*#TP
length(which(RunningData_train$NewRRI == prediction) && which(RunningData_train$NewRRI == 0))-#TN
length(which(RunningData_train$NewRRI != prediction) && which(prediction == 1))*#FP
length(which(RunningData_train$NewRRI != prediction) && which(prediction == 0)))/#FN
sqrt(
(length(which(RunningData_train$NewRRI == prediction) && which(RunningData_train$NewRRI == 1))+#TP
length(which(RunningData_train$NewRRI != prediction) && which(prediction == 1)))*#FP
(length(which(RunningData_train$NewRRI == prediction) && which(RunningData_train$NewRRI == 1))+#TP
length(which(RunningData_train$NewRRI != prediction) && which(prediction == 0)))*#FN
(length(which(RunningData_train$NewRRI == prediction) && which(RunningData_train$NewRRI == 0))+#TN
length(which(RunningData_train$NewRRI != prediction) && which(prediction == 1)))*#FP
(length(which(RunningData_train$NewRRI == prediction) && which(RunningData_train$NewRRI == 0))+#TN
length(which(RunningData_train$NewRRI != prediction) && which(prediction == 0)))#FN
)
)
}
# Inspect best MCC value:
plot(cutoffs, mcc, pch = 19, type='b',
xlab="Cutoff Level", ylab = "MCC")
# Later take the cutoff value with the best MCC on training data and use it for test set prediction:
Predictions <- RunningData_test %>%
dplyr::mutate(Prediction = predict(MLBR_L, RunningData_test, type = "response"),
Prediction = ifelse(Prediction > .5, 1, 0), ## Take the best cutoff value then
Prediction = factor(Prediction, levels = c("0", "1")),
NewRRI = factor(NewRRI, levels = c("0", "1")))
# Check MCC
mcc(RunningData_test, Predictions$NewRRI, Predictions$Prediction)
Thank you for your help in advance, kind regards!