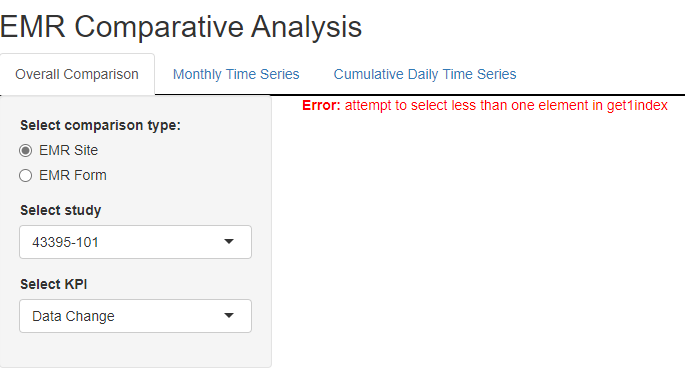
I built a simple R shiny app with multiple tabs. When I run the app, the plot on each tab would not show when I first clicked on each of the tabs. An error message shows attempt to select less than one element in get1index. However, after I clicked on each of the tabs until the last tab, plots started to show up on each tab without error message. Following is the screenshot I have when I run the app. Would you please kindly point out which direction I should pursue to solve this issue? Thanks,
Before I clicked on each of the tabs, I got the following ERROR message with no plot rendered.
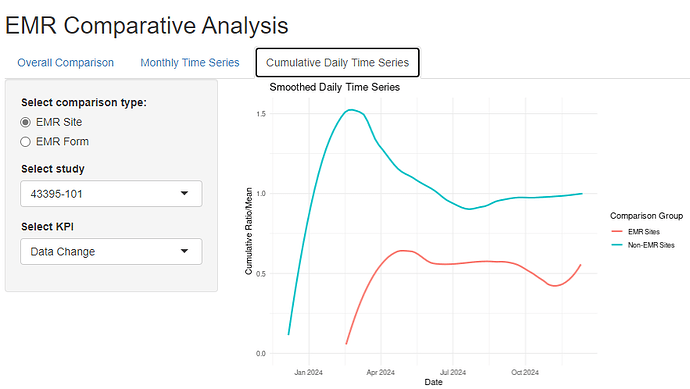
After I clicked on each of the tabs, starting from the last tab I clicked, all plots start to show up.
The simplified UI and server part is similar to the following.
ui <- fluidPage(
titlePanel("EMR Comparative Analysis"),
tabsetPanel(
tabPanel("Overall Comparison",
sidebarLayout(
sidebarPanel(
# Radio button to select between EMR Site and EMR Form
radioButtons(
inputId = "overall_comparison_type",
label = "Select comparison type:",
choices = c("EMR Site" = "emrsite", "EMR Form" = "emrform"),
selected = "emrsite"
),
# Dropdown menu for study selection
uiOutput("overallStudySelectInput"),
# Input selection for cumulative daily time series
uiOutput("overallMetricSelectInput")
),
mainPanel(
plotOutput("overallPlot")
)
)
),
}
# Server ----
server <- function(input, output, session) {
....
# Consolidate reactive expressions for data selection
selectedData <- reactive({
print("Running selectedData() reactive...")
list(
cumulative = {
# req(input$cumulative_comparison_type, input$select_cumulative_study, input$select_cumulative_metric)
print("Fetching cumulative data...")
data_list <- get(input$cumulative_comparison_type)
study_data <- data_list[[input$select_cumulative_study]]
print(paste("Selected study for cumulative TS:", input$select_cumulative_study))
kpi_data <- study_data[[input$select_cumulative_metric]]
},
monthly = {
# req(input$select_monthly_study, input$select_monthly_metric)
print("Fetching monthly data...")
data_list <- get(input$monthly_comparison_type)
study_data <- data_list[[input$select_monthly_study]]
print(paste("Selected study for monthly TS:", input$select_monthly_study))
kpi_data <- study_data[[input$select_monthly_metric]]
},
overall = {
# req(input$select_overall_study, input$select_overall_metric)
print("Fetching overall data...")
data_list <- get(input$overall_comparison_type)
study_data <- data_list[[input$select_overall_study]]
print(paste("Selected study for overall TS:", input$select_overall_study))
kpi_data <- study_data[[input$select_overall_metric]]
}
)
})
# Render overall Plot
output$overallPlot <- renderPlot({
data <- selectedData()$overall
print("overall data structure:")
print(str(data))
comparison_var <- if (input$overall_comparison_type == "emrsite") {
"is_emr_site"
} else {
"is_emr_form"
}
comparison_label <- c(
"Yes" = if (input$overall_comparison_type == "emrsite") "EMR Sites" else "EMR Forms",
"No" = if (input$overall_comparison_type == "emrsite") "Non-EMR Sites" else "Non-EMR Forms"
)
data[[comparison_var]] <- factor(data[[comparison_var]], levels = names(comparison_label), labels = comparison_label)
# Identify columns starting with 'ratio' or 'duration_median'
plot_var <- grep("^ratio|^duration_median", names(data), value = TRUE)
ggplot(data, aes_string(x = comparison_var, y = plot_var, fill = comparison_var)) +
geom_bar(stat = "identity", position = "dodge") +
labs(title = "Overall Comparison by Study", x = "EMR vs. Non-EMR", y = "Overall Ratio/Median", fill = "Comparison Group") +
theme_minimal()
})
}