Hello,
I'm coming to you with this project that is seriously poisoning my health,
I'm trying to make a shiny interactive site to present my biology internship data and it's pain in the *** continuously, this is not contractual but rather a bonus that I want to succeed as a personal project in order to progress in my professional skills
This is my first website, I already have a basic knowledge of HTML/CSS and I'm suffering, the debugging is horrible and I never understand anything I'm asked to do, unlike R markdown in particular where it's really very simple
I work on R studio Cloud with an encoding in UTF-8
Here is my code
# APPLICATION WEB PROJET M2 - PHYLOGENIE MOLECULAIRE DES BRENTIDAE ACRATINI
# LOAD DATA - - - - - - - - - - - - - - - -
# PACKAGES - - - - - - - - - - - - - - - -
library(shiny) # shiny web interactif
library(shinythemes) # theme couleur general de l'application
library(shinydashboard) # à voir mais potentiellement pas utile (visiblement cela rentre en conflit avec le code d'une app normale)
library(dendextend) # modification et contrôle des objets de type arbre ("dendrogram")
library(DT) # Datatable classe pour illustrer les datasets
# Représentation graphique
library(circlize) # pour faire des circos plot vraiment très joli
#library(ggtree) # manipulation très pratique d'arbre phylogénétique
library(ggplot2) # faire de joli plot
library(ggpubr) # pour représenter les plots côtes à côtes de manière classe
library(leaflet) # faire une très joli carte sur fond de Javascript
# SOURCES - - - - - - - - - - - - - - - -
# https://chlorobox.mpimp-golm.mpg.de/OGDraw.html => tool for plotting easy circos plot from molecular data
# CODE UI - - - - - - - - - - - - - - - -
# Define UI for application that draws a histogram
shinyUI(fluidPage(id = 'test',
tags$style('#test {
background-color: #FFFBF5;
}'),
# - - - TITRE & IMAGES DE GARDE
titlePanel(""),
HTML('<center><img src="https://www.dropbox.com/s/i6ot98mblktmcyc/Brentidae%20Acratini%20Phylogeny%20-%20Copie.png?dl=1" height="300vh"></center>'),
br(),
# - - - - NAVBAR / Onglet
navbarPage("Let's get started"),
tabPanel(icon("home"),
fluidRow(column(
tags$img(src="https://www.dropbox.com/s/mwqbiuf1yk2th8n/Carte_Amerique_Sud_avec_%20Acratini.png?dl=1",width="200px",height="300px"),width=2,
br(),
p(em("Teramocerus pulchellus Perroud 1853"),br(),"Acratini",a(href="https://www.gbif.org/fr/species/4238", "- GBIF",target="_blank"),style="text-align:center;color:black")
),
column(
br(),
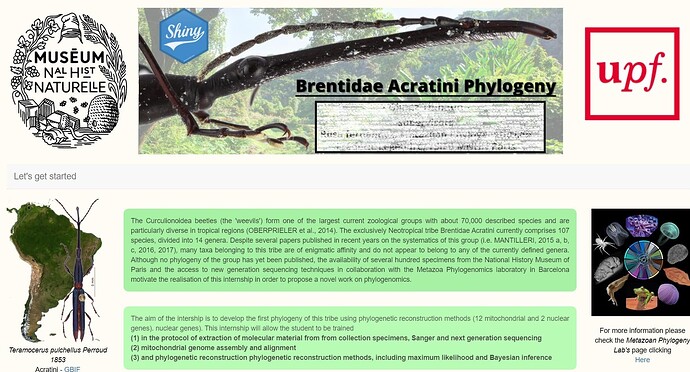
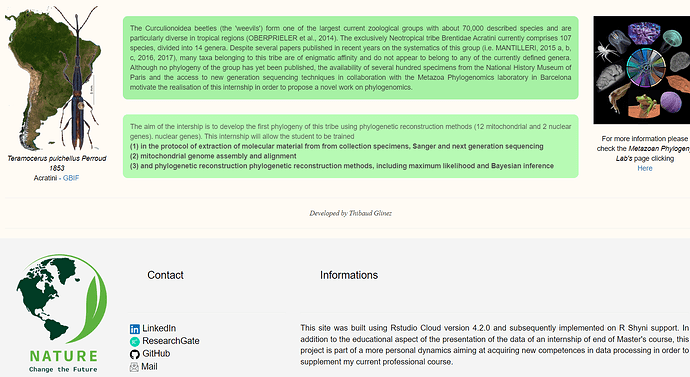
p("The Curculionoidea beetles (the 'weevils') form one of the largest current zoological groups with about 70,000
described species and are particularly diverse in tropical regions (OBERPRIELER et al., 2014). The exclusively
Neotropical tribe Brentidae Acratini currently comprises 107 species, divided into 14 genera. Despite several
papers published in recent years on the systematics of this group (i.e. MANTILLERI, 2015 a, b, c, 2016, 2017),
many taxa belonging to this tribe are of enigmatic affinity and do not appear to belong to any of the currently
defined genera. Although no phylogeny of the group has yet been published, the availability of several hundred
specimens from the National History Museum of Paris and the access to new generation sequencing
techniques in collaboration with the Metazoa Phylogenomics laboratory in Barcelona motivate the realisation of
this internship in order to propose a novel work on phylogenomics.",style="text-align:justify;color:black;background-color:lightgreen;opacity:0.8;padding:15px;border-radius:10px"),
br(),
p("The aim of the intership is to develop the first phylogeny of this tribe using phylogenetic reconstruction
methods (12 mitochondrial and 2 nuclear genes). nuclear genes). This internship will allow the student to be
trained",br(),strong("(1) in the protocol of extraction of molecular material from from collection specimens,
Sanger and next generation sequencing"),br(), strong("(2) mitochondrial genome assembly and alignment"),
br(), strong("(3) and phylogenetic reconstruction phylogenetic reconstruction methods, including maximum
likelihood and Bayesian inference"),style="text-align:justify;color:black;background-color:palegreen;opacity:0.7;padding:15px;border-radius:10px"),
width=8),
column(
br(),
tags$img(src="https://www.dropbox.com/s/b67dzexr0erhqhq/Metazoan%20Phylogeny%20Lab%20pantalla.png?dl=1",height="220px",height="130px"),
br(),
br(),
p("For more information please check the",em("Metazoan Phylogeny Lab's"),"page clicking",
br(),
a(href="https://www.ibe.upf-csic.es/metazoa-phylogenomics", "Here",target="_blank"),style="text-align:center;color:black"),
width=2)),
hr(),
p(em("Developed by Thibaud Glinez"),style="text-align:center; font-family: times"),
hr()
),
# - - - - FOOTER
fluidRow(
# - - - Left Column Brentid Logo
column(
tags$img(src="https://www.dropbox.com/s/4abpyb9g72jw3xi/Nature.png?dl=1",width="300px",height="300px"),width=2,
br()
)
# - - - Right Column Contact info
#column(
# p("oui"),
# ) # Column End
) # fluidRow End
This is what it looks like at the moment:
I'm only dealing with the UI part for the moment.
Here is my main problem, I systematically get this error:
101: )) ^ Warning: Error in sourceUTF8: Error sourcing /cloud/project/Projet_M2_Phylogenie/ui.R [No stack trace available]
Either always a problem of adding or on the contrary of removing brackets without that makes sense for the last row. So I can't add blocks in my project, either in the "NavBar" that I have to fill in or in the footer
Please could you tell me if all this is normal?
I am not an expert in R but not a beginner and this is causing me a lot of trouble
It's a bit sad from there because it really seems to be a powerful way for sharing data