I have deployed an R Shiny app at https://helmholtz-munich-zyto.shinyapps.io/LDAcoop/.
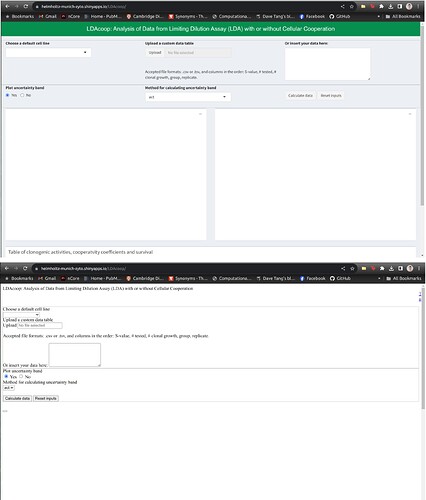
Sometimes the site loads correctly, but sometimes it appears without any colors and in a simplified form.
At the top you can see the correctly, and at bottom the incorrectly loaded versions:
Usually when it's loaded partly after a few reloads the site gets ok.
The Shiny app app.R file looks like this:
###############################################################################
##### Shiny web app of the LDAcoop library #
###############################################################################
###############################################################################
# Load libs
###############################################################################
# Load libs:
suppressPackageStartupMessages({
library("shiny")
library("LDAcoop")
library("shinyjs")
library("shinydashboard")
library("tidyverse")
library("plotly")
})
# Plotly plotting functions:
source("code/activity_SF_plot_plotly.R")
#source("/home/benedek_danko/helmholtz/projects/LDAcoop_shiny/code/activity_SF_plot_plotly.R")
###############################################################################
# Functions
###############################################################################
LDAdata <- read.csv("data/LDAdata.tsv",
sep="\t", header = T, stringsAsFactors = F)
colnames(LDAdata) <- c("name", "replicate", "Group", "S-value",
"# Tested", "# Clonal growth")
subset_data <- function(cell_line=c("MDA.MB321", "BT.20", "T47D", "HCC1806",
"SKBR3", "SKLU1", "A549", "DU4475",
"PANCO1", "PANCO2", "HNSCC.Organoid"),
data=LDAdata){
# Subsets cell line data from LDAdata
#data(LDAdata)
df_subset <- subset.data.frame(x = data,
subset = name == cell_line)
df_subset <- df_subset[,c("S-value","# Tested",
"# Clonal growth",
"Group","replicate")]
return(df_subset)
}
###############################################################################
# Global variables
###############################################################################
cell_lines <- list("",
"A549" = "a549",
"BT.20" = "bt20",
"DU4475" = "du4475",
"HCC1806" = "hcc1806",
"HNSCC.Organoid" = "hnsccorganoid",
"MDA.MB321" = "mdamb321",
"PANCO1" = "panc01",
"SKBR3"="skbr3",
"SKLU1"="sklu1",
"T47D"="t47d")
###############################################################################
# Define UI
###############################################################################
ui <- dashboardPage(
skin = "green",
dashboardHeader(title = "LDAcoop: Analysis of Data from Limiting Dilution Assay (LDA) with or without Cellular Cooperation",
titleWidth = "100%"),
dashboardSidebar(disable = TRUE),
dashboardBody(
shinyjs::useShinyjs(),
tags$head(
# links to files in www/
tags$link(rel = "stylesheet", type = "text/css", href = "basic_template.css"),
tags$link(rel = "stylesheet", type = "text/css", href = "custom.css"),
tags$script(src = "custom.js"),
tags$style(HTML('.content-wrapper { overflow: auto; }'))
),
tags$footer(
p("Maintainer(s): Daniel Samaga, Benedek Danko"),
title="Your footer here",
align = "center",
style = "
position:absolute;
bottom:0;
width:100%;
height:30px; /* Height of the footer */
color: gray;
padding: 0px;
background-color: white;
z-index: 1000;
left: 0px;"
),
# Input:
fluidRow(
style = "border: 1px solid silver;",
column(width = 4,selectInput("default_cell_lines",
label = "Choose a default cell line",
choices = cell_lines,
selected = FALSE,
multiple = FALSE)) ,
column(width = 4,fileInput("upload_file",
label = "Upload a custom data table",
multiple = FALSE,
accept = c(".csv", ".tsv"),
buttonLabel = "Upload"),
p("Accepted file formats: .csv or .tsv, and columns in the order:\nS-value, # tested, # clonal growth, group, replicate.")),
column(width = 4,textAreaInput("custom_input",
"Or insert your data here:", rows = 5))
),
# Options:
fluidRow(
style = "border: 1px solid silver;",
column(width = 4,radioButtons("uncert_band",
label = "Plot uncertainty band",
choices = c("Yes", "No"),
selected = "Yes",
inline = TRUE)),
column(width = 4, selectInput("uncert_method",
label = "Method for calculating uncertainty band",
choices = c("act"="act",
"ep"="ep"),
selected = "act",
multiple = FALSE)),
# column(width = 4,selectInput('ref','Change reference group',
# choices=NULL, selected="0")),
column(width = 4, br(), actionButton("calculate_data","Calculate data"),
actionButton("reset_input", "Reset inputs"))
),
# Plots:
fluidRow(
box(
collapsible = TRUE, title = "", solidHeader = F,
column(plotlyOutput("plt1"), width = 6, height=450)
),
box(
collapsible = TRUE, title = "", solidHeader = F,
column(plotlyOutput("plt2"), width = 6, height=450)
)
),
# Table:
box(width="100%", #background = 'green',
DT::dataTableOutput('LDA_table'),
title="Table of clonogenic activities, cooperatvity coefficients and survival",
downloadButton('export_table',"Export table")),
br(),
# Text:
p('Cellular cooperation compromises the established method of calculating clonogenic activity from limiting dilution assay (LDA) data. This tool provides functions that enable robust analysis in presence or absence of cellular cooperation. The implemented method incorporates the same cooperativity module to model the non-linearity associated with cellular cooperation as known from the colony formation assay (Brix et al. (2021) ',
a(href="https://doi.org/10.1038/s41596-021-00615-0",
"doi:10.1038/s41596-021-00615-0"),
' : "Analysis of clonogenic growth in vitro." Nature protocols).'),
br()
)
)
###############################################################################
# Define server logic
###############################################################################
server <- function(input, output, session) {
# Store and update data:
values <- reactiveValues(data=vector(mode = "list",0),
output=vector(mode = "list",0),
plot_data=vector(mode = "list",0))
# Subset data:
observeEvent(input$calculate_data, {
# Any of the inputs required:
req(
isTruthy(input$custom_input) ||
isTruthy(input$default_cell_lines) || isTruthy(input$upload_file)
)
# If user typed/pasted custom input:
if (input$custom_input!=""){
df <- do.call(rbind.data.frame,
strsplit(strsplit(input$custom_input,
"\n")[[1]],
"\\s|\\t|,|;")) %>%
mutate(across(everything(), as.character)) %>%
mutate(across(everything(), as.numeric))
colnames(df) <- c("S-value","# Tested","# Clonal growth","Group","replicate")
values$data <- df
values$plot_data <- LDA_prepare_plot(values$data,
ifelse(input$uncert_band=="Yes",
input$uncert_method,
"act"))
# If default cell line selected:
} else if (input$default_cell_lines %in% as.character(unlist(cell_lines))[-1]){
selected_cell_line <- names(cell_lines)[which(cell_lines %in% input$default_cell_lines)]
values$data <- subset_data(cell_line = selected_cell_line)
values$plot_data <- LDA_prepare_plot(values$data,
ifelse(input$uncert_band=="Yes",
input$uncert_method,
"act"))
# If input file uploaded:
} else if (input$upload_file$size > 0){
if (str_detect(input$upload_file$name, "\\.tsv$")){
file <- read.csv2(input$upload_file$datapath, sep="\t")
} else if (str_detect(input$upload_file$name, "\\.csv$")){
file <- read.csv2(input$upload_file$datapath, sep=";")
}
file <- file %>%
mutate(across(everything(), as.character)) %>%
mutate(across(everything(), as.numeric))
colnames(file) <- c("S-value","# Tested","# Clonal growth","Group","replicate")
values$data <- file
values$plot_data <- LDA_prepare_plot(values$data,
ifelse(input$uncert_band=="Yes",
input$uncert_method,
"act"))
}
}, label = "collect_cell_line_data")
# Reset:
observeEvent(input$reset_input, {
if (input$reset_input[1] > 0){
values$data <- NULL
values$output <- NULL
values$plot_data <- NULL
updateSelectInput(session=session, # reset cell line selection
inputId="default_cell_lines",
label="",
choices = cell_lines,
selected="")
reset("upload_file")} # reset file upload
})
# LDA plots:
output$plt1 <- renderPlotly(
if (length(reactiveValuesToList(values)$data) > 0){
sf_plotly(values$plot_data)
}
)
output$plt2 <- renderPlotly(
if (length(reactiveValuesToList(values)$data) > 0){
activity_plotly(values$plot_data, xlim=NULL, input$uncert_band)
}
)
# Create data table:
output$LDA_table <- DT::renderDataTable(rownames= FALSE,
{
if (length(reactiveValuesToList(values)$data) > 0){
df <- LDA_table(x = values$data, ref_class = 0,
uncertainty = input$uncert_method) %>%
mutate(act=round(act, 3),
act.CI.lb=round(act.CI.lb, 3),
act.CI.ub=round(act.CI.ub, 3),
b=round(b, 3),
b.pvalue=round(b.pvalue, 3),
SF=round(SF, 3),
SF.CI.lb=round(SF.CI.lb, 3),
SF.CI.ub=round(SF.CI.ub, 3))
values$output <- df
}
})
# Export data table:
output$export_table <- downloadHandler(
filename = function(){paste0("LDA_",
names(cell_lines)[which(cell_lines %in% input$default_cell_lines)] %>%
str_replace_all("\\.", "_"),
".csv")},
content = function(fname){
write.csv(values$output,
fname,
quote = F,
row.names = F,
sep = "\t")
})
}
###############################################################################
# Run the application
###############################################################################
shinyApp(ui = ui, server = server)
Before deployment, I installed the most recent versions of the R libraries from CRAN.
Any idea why this happens? Should I modify some settings in the site config?
Most recent logs from shinyapps.io:
2023-10-10T08:41:44.296189+00:00 shinyapps[10134802]: Starting R with process ID: '237'
2023-10-10T08:41:44.301370+00:00 shinyapps[10134802]: Shiny application starting ...
2023-10-10T08:41:46.187144+00:00 shinyapps[10134802]:
2023-10-10T08:41:46.192525+00:00 shinyapps[10134802]: Listening on http://127.0.0.1:41435
2023-10-10T08:41:55.186361+00:00 shinyapps[10134802]: R: src/unix/core.c:258: uv__finish_close: Assertion `handle->flags & UV_HANDLE_CLOSING' failed.
2023-10-10T08:41:55.194683+00:00 shinyapps[10134802]: Running on host: 179dc5ff9068
2023-10-10T08:41:55.199777+00:00 shinyapps[10134802]: Running as user: uid=10001(shiny) gid=10001(shiny) groups=10001(shiny)
2023-10-10T08:41:55.205020+00:00 shinyapps[10134802]: Connect version: 2023.03.0
2023-10-10T08:41:55.210330+00:00 shinyapps[10134802]: LANG: C.UTF-8
2023-10-10T08:41:55.215363+00:00 shinyapps[10134802]: Working directory: /srv/connect/apps/LDAcoop
2023-10-10T08:41:55.220394+00:00 shinyapps[10134802]: Using R 4.3.1
2023-10-10T08:41:55.225665+00:00 shinyapps[10134802]: R.home(): /opt/R/4.3.1/lib/R
2023-10-10T08:41:55.230677+00:00 shinyapps[10134802]: Content will use current R environment
2023-10-10T08:41:55.235883+00:00 shinyapps[10134802]: R_LIBS: (unset)
2023-10-10T08:41:55.241017+00:00 shinyapps[10134802]: .libPaths(): /opt/R/4.3.1/lib/R/library
2023-10-10T08:41:55.246939+00:00 shinyapps[10134802]: shiny version: 1.7.5
2023-10-10T08:41:55.252047+00:00 shinyapps[10134802]: httpuv version: 1.6.10
2023-10-10T08:41:55.257235+00:00 shinyapps[10134802]: rmarkdown version: 2.21
2023-10-10T08:41:55.262485+00:00 shinyapps[10134802]: knitr version: 1.42
2023-10-10T08:41:55.268402+00:00 shinyapps[10134802]: jsonlite version: 1.8.4
2023-10-10T08:41:55.273237+00:00 shinyapps[10134802]: RJSONIO version: (none)
2023-10-10T08:41:55.278191+00:00 shinyapps[10134802]: htmltools version: 0.5.5
2023-10-10T08:41:55.283226+00:00 shinyapps[10134802]: reticulate version: (none)
2023-10-10T08:41:55.288387+00:00 shinyapps[10134802]: Using pandoc: /opt/connect/ext/pandoc/2.16
2023-10-10T08:41:56.209520+00:00 shinyapps[10134802]:
2023-10-10T08:41:56.215287+00:00 shinyapps[10134802]: Starting R with process ID: '264'
2023-10-10T08:41:56.220378+00:00 shinyapps[10134802]: Shiny application starting ...
2023-10-10T08:41:58.192134+00:00 shinyapps[10134802]:
2023-10-10T08:41:58.197841+00:00 shinyapps[10134802]: Listening on http://127.0.0.1:33959