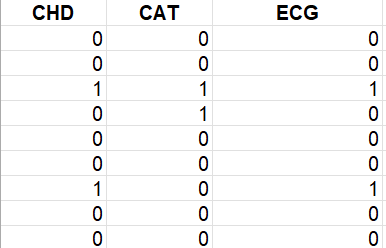
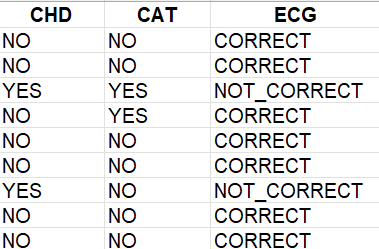
This is my dataframe and code:
bazza <- structure(list(CHD = c(0, 0, 1, 0, 0, 0, 1, 0, 0, 0, 0, 0, 0,
0, 0, 1, 0, 0, 0, 0), CAT = c(0, 0, 1, 1, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0, 1, 0, 1, 0, 0), ECG = c(0, 0, 1, 0, 0, 0, 1, 0, 0,
1, 0, 0, 0, 0, 0, 1, 0, 1, 0, 1)), class = c("tbl_df", "tbl",
"data.frame"), row.names = c(NA, -20L))
bazza$CHD <- factor(bazza$CHD, levels = c(0, 1), labels = c("NO", "YES"))
bazza$CAT <- factor(bazza$CAT, levels = c(0, 1), labels = c("NO", "YES"))
bazza$ECG <- factor(bazza$ECG, levels = c(0, 1), labels = c("NO_Pra", "YES_Niepr"))
After converting variables to factors RStudio displays labels instead of levels:

What to do to keep it like this dataframe below and still have got variables as factors ?
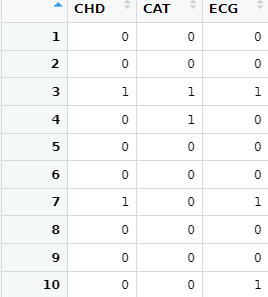
Strangely if I run:
>levels(bazza$CAT)
[1] "NO" "YES"
I set up levels as 0 and 1 not NO and YES - these are factor labels. Can someone explain that behaviour ?