Hello everyone, I hope you are wonderful.
Does anyone know why in my RDA analysis, the last variable, which is precipitation, is not being contemplated, because in the analysis range I am contemplating it (Metadata[,7:19]).
Thanks in advance
# clear my environment
rm(list=ls())
#RDA
Familia <- tibble::tribble(
~SampleID, ~Cyanobacteriaceae, ~Microcystaceae, ~Nostocaceae,
"1A", 1.342953283, 0, 0,
"1C", 1.966304234, 0, 0,
"1D", 0, 0, 1.80374141,
"1E", 1.492119211, 0, 1.492119211,
"1F", 0.597623166, 0, 0,
"1G", 1.319782597, 0, 0,
"1H", 0.859080227, 6.52598632, 0.859080227,
"2A", 0, 0, 1.907218755,
"2B", 0.793958833, 0, 0,
"2C", 0, 2.023336602, 0,
"2D", 0.859080227, 0, 0,
"2E", 0, 0, 2.333056481,
"2F", 0, 4.129704923, 0,
"2H", 0, 0, 2.333056481,
"3A", 0, 0, 2.970554964,
"3B", 0, 0, 4.592839043,
"4A", 0, 0, 0,
"4B", 0, 0, 0,
"4E", 0, 0, 0,
"4G", 0, 0, 0,
"4H", 0, 1.438454518, 3.33936827,
"5E", 0, 0, 0,
"5F", 0, 2.830692732, 0,
"5G", 0, 2.063562226, 0,
"6A", 0, 0, 2.333056481,
"6F", 0, 0, 0,
"6H", 0, 3.346178301, 0,
"7D", 0, 0, 0,
"7F", 0, 2.222744018, 0,
"7G", 0, 0, 0,
"8A", 0, 0, 0,
"8B", 0, 0, 0,
"8C", 0, 0, 0,
"8D", 0, 2.830692732, 0,
"8E", 0, 0, 0,
"8G", 0, 3.025690271, 2.155958054,
"8H", 0, 3.623754759, 0,
"9A", 0, 0, 1.80374141,
"9B", 0, 0, 1.80374141,
"9D", 0, 0, 2.333056481,
"9E", 0, 0, 0,
"9F", 0, 1.006965294, 2.333056481,
"9G", 0, 0, 0,
"9H", 0, 2.830692732, 6.06602299,
"10A", 0, 0, 0.874433896,
"10B", 0, 0, 0,
"10D", 0, 0, 0,
"10E", 0, 2.830692732, 0,
"10F", 0, 0, 0,
"10G", 0, 1.526777458, 0,
"10H", 0, 2.078731551, 0,
"11A", 0, 0, 1.80374141,
"11B", 0, 0, 1.856218562,
"11C", 0, 0, 2.333056481,
"11D", 0, 0, 2.333056481,
"11E", 0, 0, 2.333056481,
"11F", 0, 0, 0,
"11G", 0, 0, 0,
"11H", 0, 0, 0,
"12D", 0, 2.321050311, 4.034593529,
"12F", 0, 0, 0,
"12G", 0, 2.830692732, 0.238576719,
"12H", 0, 3.439569902, 4.295309968
)
attach(Familia)
Familia <- Familia[,-1]
rownames(Familia) <- SampleID
#> Warning: Setting row names on a tibble is deprecated.
#remove the rare microbes. It keeps only the microbes that are present in at least 10% of the samples
dim(Familia)
#> [1] 63 3
Familia <- Familia[,colMeans(Familia) >=.1]
dim(Familia)
#> [1] 63 3
Metadata<-tibble::tribble(
~SampleID, ~SamplingPoint, ~Depth, ~Month, ~Filter, ~SampleorReplica, ~Dissolved.oxygen, ~pH, ~WT, ~Turbidity, ~ORP, ~Ammonium, ~Nitrates, ~BGA, ~Chlrophyll, ~CE, ~AT, ~Evaporation, ~Precipitation,
"1A", "CEA1", "80 cm", "July", "20-25 µm", "Sample", 4.24, 9.94, 24.48, 87.3, 97, 4.46, 3.6, 279133, 36.6, 1.074, 13.95, 4.34, 2.55,
"1C", "CEA4", "Interstitial", "July", "20-25 µm", "Sample", 2.85, 9.98, 24.01, 98.8, 102.9, 4.37, 3.57, 265304, 40, 1.081, 13.95, 4.34, 2.55,
"1D", "CEA1", "80 cm", "August", "20-25 µm", "Sample", 3.81, 9.07, 26.67, 24.9, 94, 5.6, 3.53, 11603, 44, 1.147, 17.11, 5.8, 4.54,
"1E", "CEA3", "80 cm", "August", "20-25 µm", "Sample", 2.64, 8.99, 24.01, 43.1, 63.9, 3.38, 2.55, 218471, 35, 1.156, 17.11, 5.8, 4.54,
"1F", "CEA5", "80 cm", "August", "20-25 µm", "Sample", 4.61, 8.93, 23.5, 49.1, 64.7, 3.21, 2.28, 226658, 32.8, 1.157, 17.11, 5.8, 4.54,
"1G", "CEA2", "80 cm", "September", "20-25 µm", "Sample", 5.74, 9.24, 24.46, 68.6, 40.3, 1.93, 0.71, 267389, 44, 1.072, 17.9, 6.86, 6.88,
"1H", "CEA4", "80 cm", "September", "20-25 µm", "Sample", 3.69, 9.32, 24.65, 69.8, 48.1, 2.21, 1.2, 267144, 44.8, 1.152, 17.9, 6.86, 6.88,
"2A", "CEA1", "80 cm", "July", "0.45 µm", "Sample", 4.24, 9.94, 24.48, 87.3, 97, 4.46, 3.6, 279133, 36.6, 1.074, 13.95, 4.34, 2.55,
"2B", "CEA2", "Interstitial", "July", "20-25 µm", "Sample", 2.66, 10, 24.92, 113, 111.9, 4.75, 3.21, 270193, 39.8, 1.081, 13.95, 4.34, 2.55,
"2C", "CEA4", "Interstitial", "July", "0.45 µm", "Sample", 2.85, 9.98, 24.01, 98.8, 102.9, 4.37, 3.57, 265304, 40, 1.081, 13.95, 4.34, 2.55,
"2D", "CEA1", "80 cm", "August", "0.45 µm", "Sample", 3.81, 9.07, 26.67, 24.9, 94, 5.6, 3.53, 11603, 44, 1.147, 17.11, 5.8, 4.54,
"2E", "CEA3", "80 cm", "August", "0.45 µm", "Sample", 2.64, 8.99, 24.01, 43.1, 63.9, 3.38, 2.55, 218471, 35, 1.156, 17.11, 5.8, 4.54,
"2F", "CEA5", "80 cm", "August", "0.45 µm", "Sample", 4.61, 8.93, 23.5, 49.1, 64.7, 3.21, 2.28, 226658, 32.8, 1.157, 17.11, 5.8, 4.54,
"2H", "CEA4", "80 cm", "September", "0.45 µm", "Sample", 3.69, 9.32, 24.65, 69.8, 48.1, 2.21, 1.2, 267144, 44.8, 1.152, 17.9, 6.86, 6.88,
"3A", "CEA1", "Interstitial", "July", "20-25 µm", "Sample", 2.51, 10.04, 24.35, 87.7, 91.3, 4.5, 3.36, 267716, 39.6, 1.075, 13.95, 4.34, 2.55,
"3B", "CEA2", "Interstitial", "July", "0.45 µm", "Sample", 2.66, 10, 24.92, 113, 111.9, 4.75, 3.21, 270193, 39.8, 1.081, 13.95, 4.34, 2.55,
"4A", "CEA1", "Interstitial", "July", "0.45 µm", "Sample", 2.51, 10.04, 24.35, 87.7, 91.3, 4.5, 3.36, 267716, 39.6, 1.075, 13.95, 4.34, 2.55,
"4B", "CEA3", "80 cm", "July", "20-25 µm", "Sample", 3.04, 9.96, 24.04, 98.5, 98.9, 3.97, 4.35, 279109, 43, 1.083, 13.95, 4.34, 2.55,
"4E", "CEA3", "Interstitial", "August", "0.45 µm", "Sample", 1.83, 8.99, 23.99, 49, 64.1, 3.18, 2.44, 213497, 42.2, 1.155, 17.11, 5.8, 4.54,
"4G", "CEA2", "Interstitial", "September", "0.45 µm", "Sample", 3.02, 9.18, 24.14, 66.6, 40.6, 2.19, 0.67, 267007, 43.6, 1.061, 17.9, 6.86, 6.88,
"4H", "CEA4", "Interstitial", "September", "0.45 µm", "Sample", 3.3, 9.32, 24.67, 74.1, 49.8, 2.14, 0.87, 261363, 45.1, 1.142, 17.9, 6.86, 6.88,
"5E", "CEA3", "80 cm", "August", "0.45 µm", "Replica", 2.64, 8.99, 24.01, 43.1, 63.9, 3.38, 2.55, 218471, 35, 1.156, 17.11, 5.8, 4.54,
"5F", "CEA5", "80 cm", "August", "0.45 µm", "Replica", 4.61, 8.93, 23.5, 49.1, 64.7, 3.21, 2.28, 226658, 32.8, 1.157, 17.11, 5.8, 4.54,
"5G", "CEA2", "80 cm", "September", "0.45 µm", "Replica", 5.74, 9.24, 24.46, 68.6, 40.3, 1.93, 0.71, 267389, 44, 1.072, 17.9, 6.86, 6.88,
"6A", "CEA1", "80 cm", "July", "0.45 µm", "Replica", 4.24, 9.94, 24.48, 87.3, 97, 4.46, 3.6, 279133, 36.6, 1.074, 13.95, 4.34, 2.55,
"6F", "CEA5", "Interstitial", "August", "0.45 µm", "Replica", 1.82, 8.92, 23.43, 47.6, 60.4, 3.09, 2.06, 242331, 50.2, 1.157, 17.11, 5.8, 4.54,
"6H", "CEA4", "Interstitial", "September", "0.45 µm", "Replica", 3.3, 9.32, 24.67, 74.1, 49.8, 2.14, 0.87, 261363, 45.1, 1.142, 17.9, 6.86, 6.88,
"7D", "CEA2", "80 cm", "August", "20-25 µm", "Sample", 8.17, 9.04, 24.88, 46.9, 90.4, 2.81, 2.25, 246979, 34, 1.154, 17.11, 5.8, 4.54,
"7F", "CEA1", "80 cm", "September", "20-25 µm", "Sample", 7.69, 9.53, 26.14, 74.2, 67.7, 4.5, 1.3, 272332, 58.5, 1.072, 17.9, 6.86, 6.88,
"7G", "CEA3", "80 cm", "September", "20-25 µm", "Sample", 3.07, 9.31, 24.19, 69.9, 42, 1.99, 0.74, 269519, 44.3, 1.152, 17.9, 6.86, 6.88,
"8A", "CEA1", "Interstitial", "July", "0.45 µm", "Replica", 2.51, 10.04, 24.35, 87.7, 91.3, 4.5, 3.36, 267716, 39.6, 1.075, 13.95, 4.34, 2.55,
"8B", "CEA3", "80 cm", "July", "0.45 µm", "Replica", 3.04, 9.96, 24.04, 98.5, 98.9, 3.97, 4.35, 279109, 43, 1.083, 13.95, 4.34, 2.55,
"8C", "CEA5", "Interstitial", "July", "20-25 µm", "Sample", 2.18, 9.97, 24.4, 100.6, 102.7, 5.47, 4.11, 278137, 53.8, 1.078, 13.95, 4.34, 2.55,
"8D", "CEA2", "80 cm", "August", "0.45 µm", "Sample", 8.17, 9.04, 24.88, 46.9, 90.4, 2.81, 2.25, 246979, 34, 1.154, 17.11, 5.8, 4.54,
"8E", "CEA4", "80 cm", "August", "0.45 µm", "Sample", 2.63, 9, 24.7, 37.2, 74.9, 4.2, 2.99, 169175, 38.4, 1.179, 17.11, 5.8, 4.54,
"8G", "CEA3", "80 cm", "September", "0.45 µm", "Sample", 3.07, 9.31, 24.19, 69.9, 42, 1.99, 0.74, 269519, 44.3, 1.152, 17.9, 6.86, 6.88,
"8H", "CEA5", "80 cm", "September", "0.45 µm", "Sample", 3.7, 9.14, 22.65, 71.3, 37.1, 2.07, 0.68, 272096, 40, 1.127, 17.9, 6.86, 6.88,
"9A", "CEA2", "80 cm", "July", "20-25 µm", "Sample", 2.58, 10.06, 24.95, 95.2, 108.4, 4.62, 3.98, 279113, 38.9, 1.082, 13.95, 4.34, 2.55,
"9B", "CEA3", "Interstitial", "July", "20-25 µm", "Replica", 2.09, 9.95, 24.01, 95.9, 92.8, 4.06, 3.68, 279100, 50, 1.081, 13.95, 4.34, 2.55,
"9D", "CEA2", "Interstitial", "August", "20-25 µm", "Sample", 2.16, 8.96, 24.3, 42, 53, 3.19, 2.1, 174271, 50.3, 1.161, 17.11, 5.8, 4.54,
"9E", "CEA4", "Interstitial", "August", "20-25 µm", "Sample", 1.7, 9.02, 24.72, 36.1, 63.4, 3.97, 2.55, 159046, 55.9, 1.69, 17.11, 5.8, 4.54,
"9F", "CEA1", "Interstitial", "September", "20-25 µm", "Sample", 6.8, 9.51, 26.15, 74.4, 73.9, 3.3, 1.11, 271503, 51.3, 1.061, 17.9, 6.86, 6.88,
"9G", "CEA3", "Interstitial", "September", "20-25 µm", "Sample", 1.95, 9.39, 23.9, 44.4, 42, 3.38, 0.69, 261124, 68.2, 1.147, 17.9, 6.86, 6.88,
"9H", "CEA5", "Interstitial", "September", "20-25 µm", "Sample", 4.28, 8.85, 20.65, 71.3, 37.5, 1.18, 0.69, 271303, 34.2, 1.102, 17.9, 6.86, 6.88,
"10A", "CEA2", "80 cm", "July", "0.45 µm", "Replica", 2.58, 10.06, 24.95, 95.2, 108.4, 4.62, 3.98, 279113, 38.9, 1.082, 13.95, 4.34, 2.55,
"10B", "CEA3", "Interstitial", "July", "0.45 µm", "Replica", 2.09, 9.95, 24.01, 95.9, 92.8, 4.06, 3.68, 279100, 50, 1.081, 13.95, 4.34, 2.55,
"10D", "CEA2", "Interstitial", "August", "0.45 µm", "Sample", 2.16, 8.96, 24.3, 42, 53, 3.19, 2.1, 174271, 50.3, 1.161, 17.11, 5.8, 4.54,
"10E", "CEA4", "Interstitial", "August", "0.45 µm", "Sample", 1.7, 9.02, 24.72, 36.1, 63.4, 3.97, 2.55, 159046, 55.9, 1.69, 17.11, 5.8, 4.54,
"10F", "CEA1", "Interstitial", "September", "0.45 µm", "Sample", 6.8, 9.51, 26.15, 74.4, 73.9, 3.3, 1.11, 271503, 51.3, 1.061, 17.9, 6.86, 6.88,
"10G", "CEA3", "Interstitial", "September", "0.45 µm", "Sample", 1.95, 9.39, 23.9, 44.4, 42, 3.38, 0.69, 261124, 68.2, 1.147, 17.9, 6.86, 6.88,
"10H", "CEA5", "Interstitial", "September", "0.45 µm", "Sample", 4.28, 8.85, 20.65, 71.3, 37.5, 1.18, 0.69, 271303, 34.2, 1.102, 17.9, 6.86, 6.88,
"11A", "CEA2", "Interstitial", "July", "20-25 µm", "Replica", 2.66, 10, 24.92, 113, 111.9, 4.75, 3.21, 270193, 39.8, 1.081, 13.95, 4.34, 2.55,
"11B", "CEA4", "80 cm", "July", "20-25 µm", "Sample", 3.14, 10, 24, 96.9, 108, 4.48, 3.76, 279119, 38.5, 1.08, 13.95, 4.34, 2.55,
"11C", "CEA5", "Interstitial", "July", "20-25 µm", "Replica", 2.18, 9.97, 24.4, 100.6, 102.7, 5.47, 4.11, 278137, 53.8, 1.078, 13.95, 4.34, 2.55,
"11D", "CEA2", "80 cm", "August", "0.45 µm", "Replica", 8.17, 9.04, 24.88, 46.9, 90.4, 2.81, 2.25, 246979, 34, 1.154, 17.11, 5.8, 4.54,
"11E", "CEA4", "80 cm", "August", "0.45 µm", "Replica", 2.63, 9, 24.7, 37.2, 74.9, 4.2, 2.99, 169175, 38.4, 1.179, 17.11, 5.8, 4.54,
"11F", "CEA1", "80 cm", "September", "0.45 µm", "Replica", 7.69, 9.53, 26.14, 74.2, 67.7, 4.5, 1.3, 272332, 58.5, 1.072, 17.9, 6.86, 6.88,
"11G", "CEA3", "80 cm", "September", "0.45 µm", "Replica", 3.07, 9.31, 24.19, 69.9, 42, 1.99, 0.74, 269519, 44.3, 1.152, 17.9, 6.86, 6.88,
"11H", "CEA5", "80 cm", "September", "0.45 µm", "Replica", 3.7, 9.14, 22.65, 71.3, 37.1, 2.07, 0.68, 272096, 40, 1.127, 17.9, 6.86, 6.88,
"12D", "CEA2", "Interstitial", "August", "0.45 µm", "Replica", 2.16, 8.96, 24.3, 42, 53, 3.19, 2.1, 174271, 50.3, 1.161, 17.11, 5.8, 4.54,
"12F", "CEA1", "Interstitial", "September", "0.45 µm", "Replica", 6.8, 9.51, 26.15, 74.4, 73.9, 3.3, 1.11, 271503, 51.3, 1.061, 17.9, 6.86, 6.88,
"12G", "CEA3", "Interstitial", "September", "0.45 µm", "Replica", 1.95, 9.39, 23.9, 44.4, 42, 3.38, 0.69, 261124, 68.2, 1.147, 17.9, 6.86, 6.88,
"12H", "CEA5", "Interstitial", "September", "0.45 µm", "Replica", 4.28, 8.85, 20.65, 71.3, 37.5, 1.18, 0.69, 271303, 34.2, 1.102, 17.9, 6.86, 6.88
)
library(vegan)
#> Loading required package: permute
#> Loading required package: lattice
#> This is vegan 2.5-5
# Solo acepta variables numéricas así que debes obviar las variables de texto
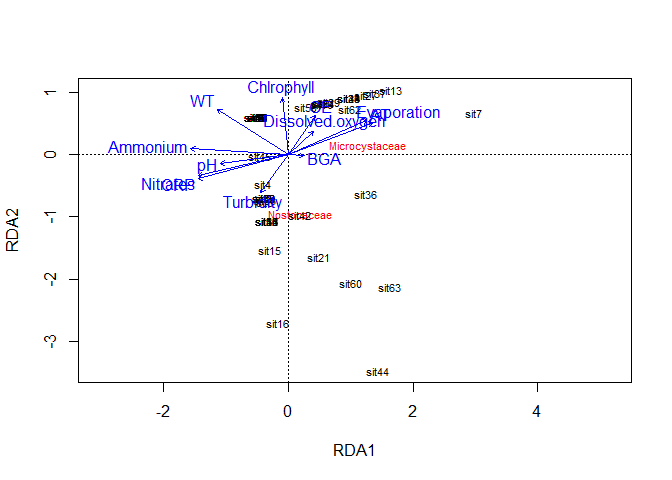
rda_object <- vegan::rda(Familia[,-1], Metadata[,7:19], scale=TRUE)
plot(rda_object)
library(BiodiversityR)
#> Loading required package: tcltk
#> Loading required package: vegan3d
#> This version of Shiny is designed to work with 'htmlwidgets' >= 1.5.
#> Please upgrade via install.packages('htmlwidgets').
#> Loading required package: rgl
#> BiodiversityR 2.11-2: Use command BiodiversityRGUI() to launch the Graphical User Interface;
#> to see changes use BiodiversityRGUI(changeLog=TRUE, backward.compatibility.messages=TRUE)
PCAsignificance(rda_object, axes=8)
#> 1 2
#> eigenvalue 0.8556295 0.6751456
#> percentage of variance 55.8951791 44.1048209
#> cumulative percentage of variance 55.8951791 100.0000000
#> broken-stick percentage 75.0000000 25.0000000
#> broken-stick cumulative % 75.0000000 100.0000000
#> % > bs% 0.0000000 1.0000000
#> cum% > bs cum% 0.0000000 0.0000000
library(devtools)
#> Loading required package: usethis
#>
#> Attaching package: 'devtools'
#> The following object is masked from 'package:permute':
#>
#> check
library(ggord)
summary(rda_object, loadings=TRUE)
#>
#> Call:
#> rda(X = Familia[, -1], Y = Metadata[, 7:19], scale = TRUE)
#>
#> Partitioning of correlations:
#> Inertia Proportion
#> Total 2.0000 1.0000
#> Constrained 0.4692 0.2346
#> Unconstrained 1.5308 0.7654
#>
#> Eigenvalues, and their contribution to the correlations
#>
#> Importance of components:
#> RDA1 RDA2 PC1 PC2
#> Eigenvalue 0.3032 0.16601 0.8556 0.6751
#> Proportion Explained 0.1516 0.08301 0.4278 0.3376
#> Cumulative Proportion 0.1516 0.23461 0.6624 1.0000
#>
#> Accumulated constrained eigenvalues
#> Importance of components:
#> RDA1 RDA2
#> Eigenvalue 0.3032 0.1660
#> Proportion Explained 0.6462 0.3538
#> Cumulative Proportion 0.6462 1.0000
#>
#> Scaling 2 for species and site scores
#> * Species are scaled proportional to eigenvalues
#> * Sites are unscaled: weighted dispersion equal on all dimensions
#> * General scaling constant of scores: 3.336994
#>
#>
#> Species scores
#>
#> RDA1 RDA2 PC1 PC2
#> Microcystaceae 1.2858 0.1385 0.8039 1.8025
#> Nostocaceae 0.1871 -0.9514 2.0292 -0.7141
#>
#>
#> Site scores (weighted sums of species scores)
#>
#> RDA1 RDA2 PC1 PC2
#> sit1 -0.5182 0.60559 -0.31478 0.193657
#> sit2 -0.5182 0.60559 -0.41778 -0.030060
#> sit3 -0.3782 -0.69443 0.13128 -0.040889
#> sit4 -0.4024 -0.46983 0.17089 -0.314519
#> sit5 -0.5182 0.60559 -0.35537 -0.332953
#> sit6 -0.5182 0.60559 -0.44806 -0.355111
#> sit7 2.9945 0.66419 0.62699 1.693459
#> sit8 -0.3702 -0.76901 0.25406 -0.060046
#> sit9 -0.5182 0.60559 -0.60514 0.291356
#> sit10 0.5502 0.81572 -0.18104 0.642632
#> sit11 -0.5182 0.60559 -0.40670 0.199049
#> sit12 -0.3372 -1.07592 0.42171 -0.426383
#> sit13 1.6625 1.03449 0.12781 1.040036
#> sit14 -0.3372 -1.07592 0.30306 -0.672284
#> sit15 -0.2877 -1.53539 0.68669 -0.402184
#> sit16 -0.1618 -2.70462 0.76471 -0.319594
#> sit17 -0.5182 0.60559 -0.19930 -0.007034
#> sit18 -0.5182 0.60559 -0.25059 -0.077542
#> sit19 -0.5182 0.60559 -0.35412 -0.089192
#> sit20 -0.5182 0.60559 -0.65337 0.017435
#> sit21 0.5005 -1.65181 0.65096 -0.275531
#> sit22 -0.5182 0.60559 -0.27414 -0.116034
#> sit23 0.9765 0.89957 -0.02418 0.608158
#> sit24 0.5715 0.81990 -0.20661 0.330954
#> sit25 -0.3372 -1.07592 0.38107 -0.116692
#> sit26 -0.5182 0.60559 -0.44488 0.035648
#> sit27 1.2487 0.95311 -0.12183 0.802933
#> sit28 -0.5182 0.60559 -0.36202 -0.168003
#> sit29 0.6555 0.83643 0.24832 0.303220
#> sit30 -0.5182 0.60559 -0.44411 -0.441234
#> sit31 -0.5182 0.60559 -0.19930 -0.007034
#> sit32 -0.5182 0.60559 -0.25059 -0.077542
#> sit33 -0.5182 0.60559 -0.29397 0.240786
#> sit34 0.9765 0.89957 -0.03083 0.773108
#> sit35 -0.5182 0.60559 -0.32221 0.045388
#> sit36 1.2468 -0.63404 0.55293 0.277916
#> sit37 1.3953 0.98194 -0.19689 0.592240
#> sit38 -0.3782 -0.69443 0.26465 -0.031609
#> sit39 -0.3782 -0.69443 0.19911 -0.143644
#> sit40 -0.3372 -1.07592 0.35466 -0.385298
#> sit41 -0.5182 0.60559 -0.23155 -0.555497
#> sit42 0.1946 -0.97134 0.55074 -0.235192
#> sit43 -0.5182 0.60559 -0.21058 -0.329458
#> sit44 1.4473 -3.47240 1.04843 -0.484502
#> sit45 -0.4504 -0.02464 -0.01252 0.092010
#> sit46 -0.5182 0.60559 -0.33887 0.096294
#> sit47 -0.5182 0.60559 -0.34119 -0.074949
#> sit48 0.9765 0.89957 0.09965 0.385613
#> sit49 -0.5182 0.60559 -0.26293 -0.259626
#> sit50 0.2880 0.76415 -0.03194 0.178144
#> sit51 0.5795 0.82148 -0.84878 0.072412
#> sit52 -0.3782 -0.69443 -0.06716 0.051418
#> sit53 -0.3742 -0.73225 0.17886 -0.265612
#> sit54 -0.3372 -1.07592 0.40188 -0.069562
#> sit55 -0.3372 -1.07592 0.33383 -0.478351
#> sit56 -0.3372 -1.07592 0.37364 -0.264961
#> sit57 -0.5182 0.60559 -0.01175 -0.435767
#> sit58 -0.5182 0.60559 -0.44411 -0.441234
#> sit59 -0.5182 0.60559 -0.62088 -0.612537
#> sit60 1.0205 -2.06121 1.13372 0.160031
#> sit61 -0.5182 0.60559 -0.26293 -0.259626
#> sit62 0.9951 0.72762 0.19178 0.579916
#> sit63 1.6314 -2.13295 0.59154 -0.046527
#>
#>
#> Site constraints (linear combinations of constraining variables)
#>
#> RDA1 RDA2 PC1 PC2
#> con1 -0.505223 -0.2085999 -0.31478 0.193657
#> con2 -0.129575 -0.1864869 -0.41778 -0.030060
#> con3 -0.435242 -0.3949468 0.13128 -0.040889
#> con4 -0.138605 0.1826363 0.17089 -0.314519
#> con5 0.210023 0.2407644 -0.35537 -0.332953
#> con6 0.316282 0.0806129 -0.44806 -0.355111
#> con7 0.278841 0.1962513 0.62699 1.693459
#> con8 -0.505223 -0.2085999 0.25406 -0.060046
#> con9 -0.388529 -0.8782862 -0.60514 0.291356
#> con10 -0.129575 -0.1864869 -0.18104 0.642632
#> con11 -0.435242 -0.3949468 -0.40670 0.199049
#> con12 -0.138605 0.1826363 0.42171 -0.426383
#> con13 0.210023 0.2407644 0.12781 1.040036
#> con14 0.278841 0.1962513 0.30306 -0.672284
#> con15 -0.342271 0.2203889 0.68669 -0.402184
#> con16 -0.388529 -0.8782862 0.76471 -0.319594
#> con17 -0.342271 0.2203889 -0.19930 -0.007034
#> con18 -0.208119 0.1903086 -0.25059 -0.077542
#> con19 -0.106376 -0.0017517 -0.35412 -0.089192
#> con20 0.006212 -0.6979111 -0.65337 0.017435
#> con21 0.312003 -0.0936548 0.65096 -0.275531
#> con22 -0.138605 0.1826363 -0.27414 -0.116034
#> con23 0.210023 0.2407644 -0.02418 0.608158
#> con24 0.316282 0.0806129 -0.20661 0.330954
#> con25 -0.505223 -0.2085999 0.38107 -0.116692
#> con26 -0.191887 -0.3058754 -0.44488 0.035648
#> con27 0.312003 -0.0936548 -0.12183 0.802933
#> con28 0.002199 0.0618938 -0.36202 -0.168003
#> con29 0.055352 1.0204349 0.24832 0.303220
#> con30 0.424393 0.1749367 -0.44411 -0.441234
#> con31 -0.342271 0.2203889 -0.19930 -0.007034
#> con32 -0.208119 0.1903086 -0.25059 -0.077542
#> con33 -0.583613 -0.2150132 -0.29397 0.240786
#> con34 0.002199 0.0618938 -0.03083 0.773108
#> con35 -0.307188 -0.0742157 -0.32221 0.045388
#> con36 0.424393 0.1749367 0.55293 0.277916
#> con37 0.793989 -0.0008145 -0.19689 0.592240
#> con38 -0.558905 -0.1417627 0.26465 -0.031609
#> con39 -0.359096 -0.1581716 0.19911 -0.143644
#> con40 -0.135623 0.0093748 0.35466 -0.385298
#> con41 0.394260 0.7081451 -0.23155 -0.555497
#> con42 0.037767 0.3490231 0.55074 -0.235192
#> con43 0.084282 0.5222431 -0.21058 -0.329458
#> con44 1.196374 -0.9218995 1.04843 -0.484502
#> con45 -0.558905 -0.1417627 -0.01252 0.092010
#> con46 -0.359096 -0.1581716 -0.33887 0.096294
#> con47 -0.135623 0.0093748 -0.34119 -0.074949
#> con48 0.394260 0.7081451 0.09965 0.385613
#> con49 0.037767 0.3490231 -0.26293 -0.259626
#> con50 0.084282 0.5222431 -0.03194 0.178144
#> con51 1.196374 -0.9218995 -0.84878 0.072412
#> con52 -0.388529 -0.8782862 -0.06716 0.051418
#> con53 -0.180285 -0.1132616 0.17886 -0.265612
#> con54 -0.583613 -0.2150132 0.40188 -0.069562
#> con55 0.002199 0.0618938 0.33383 -0.478351
#> con56 -0.307188 -0.0742157 0.37364 -0.264961
#> con57 0.055352 1.0204349 -0.01175 -0.435767
#> con58 0.424393 0.1749367 -0.44411 -0.441234
#> con59 0.793989 -0.0008145 -0.62088 -0.612537
#> con60 -0.135623 0.0093748 1.13372 0.160031
#> con61 0.037767 0.3490231 -0.26293 -0.259626
#> con62 0.084282 0.5222431 0.19178 0.579916
#> con63 1.196374 -0.9218995 0.59154 -0.046527
#>
#>
#> Biplot scores for constraining variables
#>
#> RDA1 RDA2 PC1 PC2
#> Dissolved.oxygen 0.22068 0.20399 0 0
#> pH -0.59164 -0.08247 0 0
#> WT -0.61565 0.39028 0 0
#> Turbidity -0.24107 -0.33520 0 0
#> ORP -0.78535 -0.21349 0 0
#> Ammonium -0.84956 0.04916 0 0
#> Nitrates -0.78493 -0.18740 0 0
#> BGA 0.13815 -0.01239 0 0
#> Chlrophyll -0.05463 0.49500 0 0
#> CE 0.23360 0.33560 0 0
#> AT 0.68488 0.29878 0 0
#> Evaporation 0.73787 0.27810 0 0
levels(Metadata$Month) <- c("Jul","Aug","Sep")
Familia <- Metadata$Month
bg <- c("#ff7f00","#1f78b4","#ffff33","#a6cee3","#33a02c","#e31a1c") # 6 nice colors for our ecotypes
levels(Metadata$Month) <- c("Jul","Aug","Sep")
Familia <- Metadata$Month
bg <- c("#ff7f00","#1f78b4","#ffff33","#a6cee3","#33a02c","#e31a1c") # 6 nice colors for our ecotypes
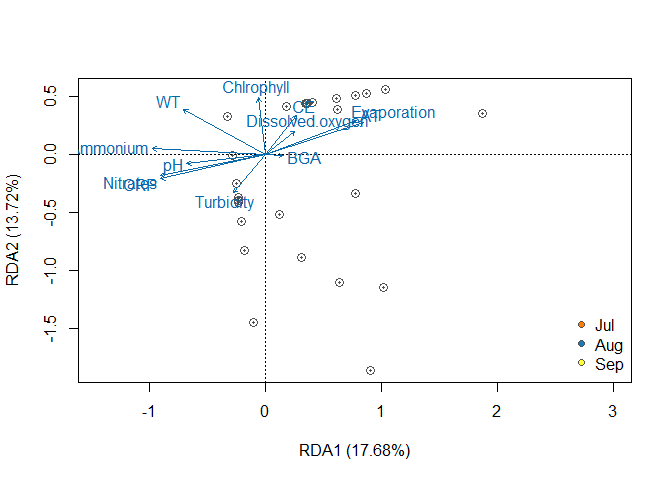
# axes 1 & 2
plot(rda_object, type = "n", scaling=3, display = "sites", xlab = "RDA1 (17.68%)", ylab = "RDA2 (13.72%)")
points(rda_object, display="sites", pch=20, cex=0.7, col="gray32", scaling=3)
points(rda_object, display="sites", pch=21, cex=1.3, col="gray32", scaling=3, bg=bg[Familia])
text(rda_object, scaling=3, display="bp", col="#0868ac", cex=1, face ="bold")
#> Warning in arrows(0, 0, pts[, 1], pts[, 2], length = head.arrow, ...):
#> "face" is not a graphical parameter
#> Warning in strwidth(labels, ...): "face" is not a graphical parameter
#> Warning in strheight(labels, ...): "face" is not a graphical parameter
#> Warning in text.default(pts, labels = rownames(pts), ...): "face" is not a
#> graphical parameter
legend("bottomright", legend=levels(Familia), bty="n",xpd= FALSE, col="gray32", pch=21, cex=1, pt.bg=bg)
Created on 2020-02-07 by the reprex package (v0.3.0)