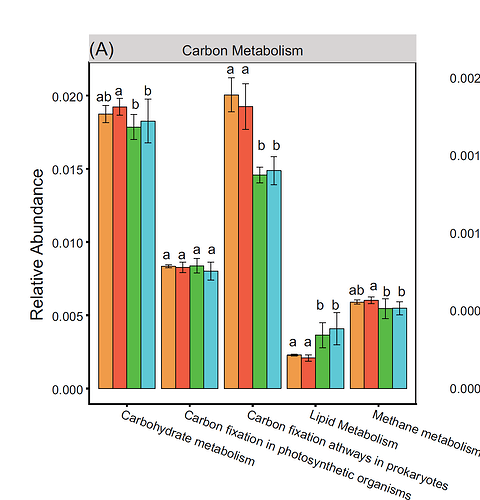
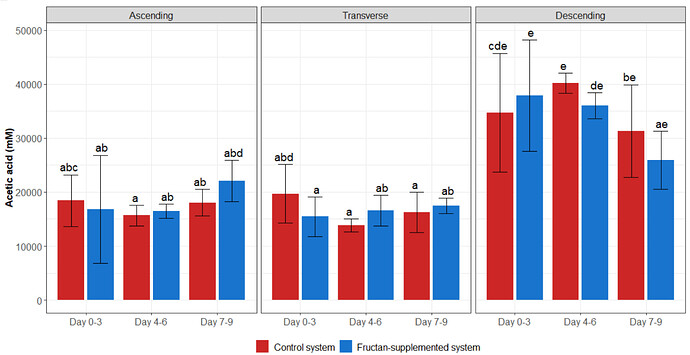
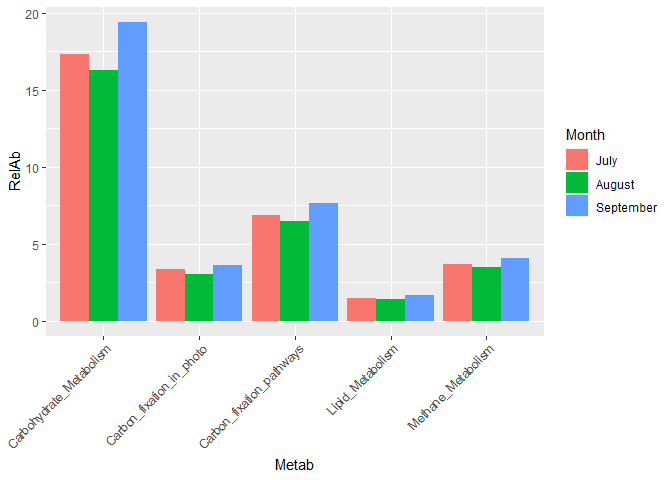
Hello to everyone. I have many weeks struggling, in how to create a graphic identical to this one. Different colors means, different months.
I have many example scripts, which I think could work for me, however I can't understand how to make the graph because I don't understand much the R language.
First I attach my database that I want to graph.
metabolism <- data.frame (tibble::tribble(
~index, ~Lipid_Metabolism, ~Methane_Metabolism, ~Carbon_fixation_pathways, ~Carbon_fixation_in_photo, ~Carbohydrate_Metabolism,
"1A", 100952L, 205749L, 427526L, 194183L, 1102658L,
"1B", 43471L, 96483L, 184384L, 92951L, 460679L,
"1C", 34365L, 95124L, 156089L, 80221L, 414818L,
"1D", 61363L, 123254L, 274105L, 108874L, 727106L,
"1E", 89307L, 219184L, 398861L, 172597L, 969309L,
"1F", 115759L, 321731L, 576492L, 263299L, 1449017L,
"1G", 25613L, 66318L, 122026L, 62049L, 298721L,
"1H", 102028L, 243559L, 456062L, 226173L, 1138806L,
"2A", 139928L, 378966L, 697790L, 344776L, 1707422L,
"2B", 87790L, 240425L, 435712L, 225590L, 1036352L,
"2C", 106988L, 270626L, 499418L, 265303L, 1245350L,
"2D", 53657L, 128471L, 234452L, 111302L, 601111L,
"2E", 104639L, 249852L, 491777L, 233460L, 1251090L,
"2F", 55292L, 137511L, 251460L, 130204L, 636126L,
"2G", 130006L, 338138L, 600900L, 315316L, 1550773L,
"2H", 124033L, 321381L, 575905L, 274012L, 1359389L,
"3A", 85026L, 229433L, 402362L, 195089L, 1023811L,
"3B", 4950L, 15343L, 26968L, 15774L, 69027L,
"3C", 24689L, 62234L, 112708L, 60231L, 283953L,
"3D", 25273L, 58473L, 123968L, 57091L, 320182L,
"3E", 40429L, 102372L, 190008L, 86958L, 457311L,
"3F", 35843L, 92503L, 169113L, 84153L, 427877L,
"3G", 34830L, 98523L, 175562L, 81875L, 444863L,
"3H", 5769L, 12359L, 24425L, 11863L, 62544L,
"4A", 201436L, 599870L, 1072543L, 483801L, 2573658L,
"4B", 92949L, 234600L, 437080L, 225848L, 1081005L,
"4C", 37854L, 95268L, 176045L, 94274L, 433309L,
"4D", 66068L, 170391L, 311889L, 148161L, 779519L,
"4E", 67192L, 189600L, 341740L, 158470L, 849012L,
"4F", 4639L, 12051L, 27180L, 12316L, 72374L,
"4G", 120771L, 306709L, 561477L, 271004L, 1458709L,
"4H", 334172L, 758954L, 1450249L, 656117L, 3632062L,
"5A", 96966L, 219273L, 440273L, 196075L, 1188916L,
"5B", 37336L, 101553L, 181427L, 91029L, 471876L,
"5C", 19846L, 44571L, 85424L, 42647L, 227428L,
"5D", 60323L, 150931L, 279470L, 141683L, 689491L,
"5E", 97331L, 248995L, 442073L, 232602L, 1140031L,
"5F", 123196L, 276931L, 531267L, 246651L, 1335246L,
"5G", 58895L, 136787L, 255109L, 103688L, 660650L,
"5H", 81282L, 183059L, 370237L, 173629L, 982498L,
"6A", 174984L, 461199L, 846430L, 411433L, 2093964L,
"6B", 167765L, 392453L, 752015L, 367876L, 1893403L,
"6C", 41837L, 111749L, 206425L, 103190L, 511093L,
"6D", 61468L, 144520L, 268953L, 140832L, 674653L,
"6E", 51700L, 146276L, 260444L, 117539L, 652398L,
"6F", 92422L, 250250L, 451339L, 214969L, 1116960L,
"6G", 48481L, 129943L, 237650L, 115942L, 654239L,
"6H", 262190L, 664679L, 1214642L, 634204L, 3079128L,
"7A", 159235L, 320985L, 667514L, 280521L, 1758378L,
"7B", 27706L, 78108L, 133526L, 67531L, 356261L,
"7C", 49006L, 132654L, 231523L, 128231L, 612022L,
"7D", 149066L, 371236L, 690310L, 378432L, 1735005L,
"7E", 129097L, 306179L, 577461L, 269750L, 1456519L,
"7F", 150763L, 317120L, 617753L, 288016L, 1595046L,
"7G", 24908L, 66217L, 118907L, 42893L, 294237L,
"7H", 24121L, 55129L, 112820L, 50952L, 300317L,
"8A", 138540L, 351913L, 658568L, 324117L, 1622058L,
"8B", 204645L, 531841L, 969287L, 469781L, 2425227L,
"8C", 57998L, 142770L, 268074L, 147259L, 662924L,
"8D", 118812L, 338736L, 598925L, 270389L, 1517644L,
"8E", 74968L, 194744L, 362079L, 158512L, 879297L,
"8F", 210033L, 483247L, 911678L, 423395L, 2308067L,
"8G", 125849L, 294075L, 552272L, 219120L, 1376611L,
"8H", 168533L, 356341L, 691371L, 311040L, 1773099L,
"9A", 39038L, 108716L, 193626L, 95556L, 519894L,
"9B", 87174L, 224827L, 403645L, 204987L, 1038656L,
"9C", 28823L, 62251L, 128463L, 55045L, 332038L,
"9D", 144455L, 409314L, 734859L, 327915L, 1849618L,
"9E", 261169L, 543774L, 1067353L, 485929L, 2708030L,
"9F", 207646L, 520002L, 957885L, 508302L, 2384944L,
"9G", 66474L, 137229L, 292459L, 120848L, 807886L,
"9H", 31011L, 102698L, 169538L, 96131L, 429061L,
"10A", 80713L, 213068L, 391096L, 199166L, 977134L,
"10B", 90113L, 219030L, 412292L, 221008L, 1023166L,
"10C", 28831L, 69206L, 126540L, 60583L, 325017L,
"10D", 72261L, 181370L, 337523L, 192536L, 878201L,
"10E", 55320L, 151178L, 267751L, 125241L, 663980L,
"10F", 75816L, 193460L, 359962L, 185979L, 876466L,
"10G", 26193L, 60943L, 116610L, 51587L, 292457L,
"10H", 26620L, 61586L, 117182L, 58476L, 300703L,
"11A", 9720L, 22885L, 51784L, 21941L, 130836L,
"11B", 8164L, 19673L, 42474L, 20125L, 107868L,
"11C", 59113L, 126480L, 252531L, 115365L, 664767L,
"11D", 82252L, 238039L, 424584L, 190519L, 1080340L,
"11E", 82156L, 200653L, 377830L, 181659L, 920255L,
"11F", 40681L, 102620L, 186620L, 95502L, 488662L,
"11G", 44678L, 87211L, 198732L, 79928L, 537708L,
"11H", 98237L, 252856L, 444224L, 239157L, 1175974L,
"12A", 164124L, 399083L, 753174L, 409195L, 1859773L,
"12B", 82461L, 204662L, 378959L, 188858L, 925236L,
"12C", 102811L, 249939L, 466436L, 254780L, 1153474L,
"12D", 118268L, 284315L, 522311L, 202894L, 1301240L,
"12E", 73010L, 164720L, 316686L, 155240L, 810844L,
"12F", 106812L, 282601L, 516927L, 246928L, 1294322L,
"12G", 283777L, 655163L, 1219255L, 530847L, 3065082L,
"12H", 208530L, 547407L, 976464L, 523509L, 2530333L
)
)
Metadata <- data.frame(tibble::tribble(
~SampleID, ~Month,
"1A", "July",
"1B", "July",
"1C", "July",
"1D", "August",
"1E", "August",
"1F", "August",
"1G", "September",
"1H", "September",
"2A", "July",
"2B", "July",
"2C", "July",
"2D", "August",
"2E", "August",
"2F", "August",
"2G", "September",
"2H", "September",
"3A", "July",
"3B", "July",
"3C", "July",
"3D", "August",
"3E", "August",
"3F", "August",
"3G", "September",
"3H", "September",
"4A", "July",
"4B", "July",
"4C", "July",
"4D", "August",
"4E", "August",
"4F", "August",
"4G", "September",
"4H", "September",
"5A", "July",
"5B", "July",
"5C", "July",
"5D", "August",
"5E", "August",
"5F", "August",
"5G", "September",
"5H", "September",
"6A", "July",
"6B", "July",
"6C", "July",
"6D", "August",
"6E", "August",
"6F", "August",
"6G", "September",
"6H", "September",
"7A", "July",
"7B", "July",
"7C", "July",
"7D", "August",
"7E", "August",
"7F", "September",
"7G", "September",
"7H", "September",
"8A", "July",
"8B", "July",
"8C", "July",
"8D", "August",
"8E", "August",
"8F", "September",
"8G", "September",
"8H", "September",
"9A", "July",
"9B", "July",
"9C", "July",
"9D", "August",
"9E", "August",
"9F", "September",
"9G", "September",
"9H", "September",
"10A", "July",
"10B", "July",
"10C", "July",
"10D", "August",
"10E", "August",
"10F", "September",
"10G", "September",
"10H", "September",
"11A", "July",
"11B", "July",
"11C", "July",
"11D", "August",
"11E", "August",
"11F", "September",
"11G", "September",
"11H", "September",
"12A", "July",
"12B", "July",
"12C", "August",
"12D", "August",
"12E", "August",
"12F", "September",
"12G", "September",
"12H", "September"
)
)
Created on 2021-07-05 by the reprex package (v0.3.0)
Created on 2021-07-05 by the reprex package (v0.3.0)
``
In this section, it is an example that I found on the internet where they get this :
It's different, but I think this script can help me
#Function bar graphics
bargraf2 <- function(df, ac, labs){
CDT2 <- c("firebrick3", "dodgerblue3")
x_lab <- rep(seq(0.75,3.25, by=0.5), 3)
ylab1 <- df$media+df$dest
y_lab <- c(ylab1[1],ylab1[4], ylab1[2], ylab1[5], ylab1[3], ylab1[6],
ylab1[7],ylab1[10], ylab1[8], ylab1[11], ylab1[9], ylab1[12],
ylab1[13],ylab1[16], ylab1[14], ylab1[17], ylab1[15], ylab1[18])
ggplot(df)+
geom_bar(aes(x=Day, y=media, fill=tr),stat = "identity", position = "dodge2")+
geom_errorbar(aes(x=Day, y=media, ymin=if_else( (media-dest)>=0, media-dest, 0 ), ymax=media+dest), position=position_dodge2(width=0.2, padding=0.5))+
labs(x=NULL, y= paste(ac, "(mM)"))+
facet_wrap(~co)+
scale_fill_manual(values = CDT2)+
geom_text(aes(x=x_lab,y=y_lab+min(media)*0.05,label=labs), size=5, vjust="bottom")+
#geom_text(aes(x=x_lab,y=media+min(media)*0.05,label=labs), size=5, vjust="bottom")+
theme_bw()+
theme(
legend.position="bottom",
legend.title = element_blank(),
legend.text = element_text(size=12),
axis.text.x=element_text(size=12),
axis.text.y=element_text(size=12),
axis.title.x = element_text(face="bold", size=13),
axis.title.y = element_text(face="bold", size=13),
strip.text.x = element_text(size = 12)
)
}
#Metabolism
AA_val <- dplyr::select(total, AA, co, Day_tr) %>% group_by(Day_tr, co) %>% summarise(media=mean(AA), dest= sd(AA)) %>% ungroup()
#> Error in dplyr::select(total, AA, co, Day_tr) %>% group_by(Day_tr, co) %>% : no se pudo encontrar la función "%>%"
#acids_gr <- total %>% group_by(Day_tr, co)
bar_anova_gr <- aov(AA ~ Day_tr_co, data=total, na.action=na.omit)
#> Error in terms.formula(formula, "Error", data = data): objeto 'total' no encontrado
Pairs_bar_gr <- glht(bar_anova_gr, linfct = mcp(Day_tr_co = "Tukey"))
#> Error in glht(bar_anova_gr, linfct = mcp(Day_tr_co = "Tukey")): no se pudo encontrar la función "glht"
lab_bar_gr <- cld(Pairs_bar_gr)$mcletters$Letters %>% as.vector()
#> Error in cld(Pairs_bar_gr)$mcletters$Letters %>% as.vector(): no se pudo encontrar la función "%>%"
AA_val_2 <- dplyr::select(total, AA, co, tr, Day, Day_tr) %>% group_by(co, tr, Day, Day_tr) %>% summarise(media=mean(AA), dest= sd(AA)) %>% ungroup()
#> Error in dplyr::select(total, AA, co, tr, Day, Day_tr) %>% group_by(co, : no se pudo encontrar la función "%>%"
This is the imagene that obtained with the previous script:
Created on 2021-07-05 by the reprex package (v0.3.0)
As you could see, the initial graph is of relative abundance. The most I could get with all the resources from different packages was the following. However, it is not very close to the final graphic that I would like to obtain (first image)
library(readxl)
Genero <- read_excel("~/RSTUDIOPICRUST/Carbono/Barplot_Carbono.xlsx")
data<- Genero
attach(Genero)
rwnames <- index
data <- as.data.frame(data[,-1])
rownames(data) <- rwnames
Metadata<- read.csv("~/RSTUDIOPICRUST/Metadata-completa.csv", row.names=1)
#SUBSET DE MONTH
Jul <- subset(data, Metadata$Month == "July", select = c(`Lipid_Metabolism`:`Carbohydrate_Metabolism`))
Aug <- subset(data, Metadata$Month == "August", select = c(`Lipid_Metabolism`:`Carbohydrate_Metabolism`))
Sep <- subset(data, Metadata$Month == "September", select = c(`Lipid_Metabolism`:`Carbohydrate_Metabolism`))
Metadata$Month <- factor(Metadata$Month,
levels = c("Jul", "Aug", "Sep"))
#July
Jul <- data.frame(Jul)
Jul_counts <- colSums(Jul)
Counts <- unname(Jul_counts)
Jul_counts <- data.frame(Jul_counts)
Jul_counts <- t(Jul_counts)
total <- sum(Counts)
rel_ab <- Jul_counts/total
Others <- rel_ab[,colMeans(rel_ab)<.00]
Others <- sum(Others)
rel_ab <- rel_ab[,colMeans(rel_ab)>=.00]
rel_ab <- data.frame(t(rel_ab), Others)
rel_ab_P <- t(rel_ab)
abundance <- c("abundance")
rel_ab_P <- data.frame(rel_ab_P)
write.csv(rel_ab_P, file = "~/RSTUDIOPICRUST/Carbono/JUL.csv")
Jul <- read.csv("~/RSTUDIOPICRUST/Carbono/JUL.csv")
#August
Aug <- data.frame(Aug)
Aug_counts <- colSums(Aug)
Counts <- unname(Aug_counts)
Aug_counts <- data.frame(Aug_counts)
Aug_counts <- t(Aug_counts)
total <- sum(Counts)
rel_ab <- Aug_counts/total
Others <- rel_ab[,colMeans(rel_ab)<.00]
Others <- sum(Others)
rel_ab <- rel_ab[,colMeans(rel_ab)>=.00]
rel_ab <- data.frame(t(rel_ab), Others)
rel_ab_P <- t(rel_ab)
abundance <- c("abundance")
rel_ab_P <- data.frame(rel_ab_P)
write.csv(rel_ab_P, file = "~/RSTUDIOPICRUST/Carbono/AUG.csv")
Aug <- read.csv("~/RSTUDIOPICRUST/Carbono/AUG.csv")
#September
Sep <- data.frame(Sep)
Sep_counts <- colSums(Sep)
Counts <- unname(Sep_counts)
Sep_counts <- data.frame(Sep_counts)
Sep_counts <- t(Sep_counts)
total <- sum(Counts)
rel_ab <- Sep_counts/total
Others <- rel_ab[,colMeans(rel_ab)<.00]
Others <- sum(Others)
rel_ab <- rel_ab[,colMeans(rel_ab)>=.00]
rel_ab <- data.frame(t(rel_ab), Others)
rel_ab_P <- t(rel_ab)
abundance <- c("abundance")
rel_ab_P <- data.frame(rel_ab_P)
write.csv(rel_ab_P, file = "~/RSTUDIOPICRUST/Carbono/SEP.csv")
Sep <- read.csv("~/RSTUDIOPICRUST/Carbono/SEP.csv")
Family_colors <- c(
"#0048BA", "#B0BF1A",
"#7CB9E8",
"#C0E8D5",
"#B284BE",
"#72A0C1",
"#EDEAE0",
"#C46210",
"#CD9575",
"#E52B50",
"#9F2B68",
"#F19CBB",
"#848482",
"#BCD4E6",
"#9F8170",
"#3D2B1F",
"#967117",
"#CAE00D",
"#7BB661",
"#91A3B0"
)
library(ggplot2)
library(scales)
ggplot() +geom_bar(aes(y = rel_ab_P*100, x= "Jul", fill = X), data = Jul,
stat="identity", width = .5)+ geom_bar(aes(y = rel_ab_P*100, x= "Aug", fill = X), data = Aug,
stat="identity",width=.5)+
scale_x_discrete(
labels = c("Jul", "Aug", "Sep"),
drop = FALSE
) +
geom_bar(aes(y = rel_ab_P*100, x= "Sep", fill = X), data = Sep,
stat="identity", width = .5)+
theme_classic()+
theme(legend.title = element_blank())+
ylab("Relative Abundance >.01% \n")+
xlab("Month")+
scale_fill_manual(values = Family_colors)

Created on 2021-07-05 by the reprex package (v0.3.0)
The database used for the last example is the same one that I attached at the beginning of this topic.