Hi,
I'm trying to learn how to use multiple model with tune and workflow_set.
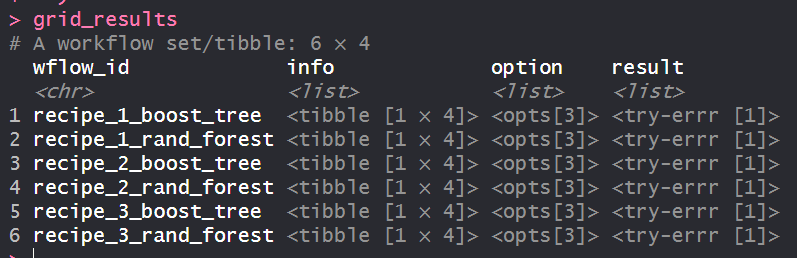
When I try apply workflow_map to my workflow_set using my CV resamples I get no error however when I inspect the grid results I see the results column contains 'try-errr'.
I'm trying to follow along to this example in the TMwR book using the ames dataset, my code looks as follows:
pacman::p_load(tidymodels,tidyverse,doParallel,janitor,AmesHousing,vip)
set.seed(1234)
# load the housing data and clean names
ames_data <- make_ames() %>%
janitor::clean_names()
# split into training and testing datasets. Stratify by Sale price
ames_split <- rsample::initial_split(
ames_data,
prop = 0.8,
strata = sale_price
)
# CREATE TRAINING AND TESTING OBJECTS FROM THE SPLIT OBJECT
ames_train <- training(ames_split)
ames_test <- testing(ames_split)
# CREATE RESAMPLES TO CHOOSE AND COMPARE MODELS
set.seed(234)
ames_folds <- vfold_cv(ames_train, strata = sale_price)
# EDA ---------------------------------------------------------------------
# DEFINE PREPROCESSING RECIPES --------------------------------------------
base_rec <-
recipe(sale_price ~ ., data = ames_train) %>%
step_log(sale_price, base = 10) %>%
step_YeoJohnson(lot_area, gr_liv_area) %>%
step_other(neighborhood, threshold = .1) %>%
step_dummy(all_nominal()) %>%
step_zv(all_predictors()) %>%
step_ns(longitude, deg_free = tune("long df")) %>%
step_ns(latitude, deg_free = tune("lat df"))
# REDUCE THE CARDINALITY OF SOME OF THE CATEGORICAL VARIABLES
low_cardinality_recipe <- base_rec %>%
# convert categorical variables to factors
recipes::step_string2factor(all_nominal()) %>%
# combine low frequency factor levels
recipes::step_other(all_nominal(), other = "Other", threshold = 0.01) %>%
# remove no variance predictors which provide no predictive information
recipes::step_nzv(all_nominal())
# CREATE A PRINCIPAL COMPONENT ANALYSIS RECIPE
pca_recipe <- recipe(sale_price ~ ., data = ames_train) %>%
step_normalize(all_numeric_predictors()) %>%
step_pca(all_numeric_predictors(), num_comp = tune())
# # UPDATE THE GENERIC PARAMETERS FOR DEGREES OF FREEDOM SINCE IT HAS A SMALL RANGE
# # SPLINE DEGREES HAS MORE APPROPIATE VALUES FOR SPLINES
# ames_param <-
# base_rec %>%
# parameters() %>%
# update(
# `long df` = spline_degree(),
# `lat df` = spline_degree()
# )
# BUILD A MODEL -----------------------------------------------------------
# DEFINE AN XGBOOST MODEL
xgb_spec <- boost_tree(
trees = 500,
tree_depth = tune(),
min_n = tune(),
loss_reduction = tune(),
sample_size = tune(),
mtry = tune(),
learn_rate = tune(),
engine = "xgboost",
mode = "regression"
)
# DEFINE AN RANDOM FOREST MODEL
rf_spec <- rand_forest(
trees = 500,
mtry = tune(),
min_n = tune(),
engine = "ranger",
mode = "regression"
)
# DEFINE A WORKFLOW SET ---------------------------------------------------
ames_set <- workflowsets::workflow_set(
preproc = list(base_rec, low_cardinality_recipe, pca_recipe),
models = list(xgb_spec, rf_spec)
)
ames_set
# SET UP PARALLEL PROCESSING
doParallel::registerDoParallel(detectCores())
grid_ctrl <-
control_grid(
save_pred = TRUE,
parallel_over = "everything",
save_workflow = TRUE
)
grid_results <- ames_set %>%
workflow_map(
seed = 1234,
resamples = ames_folds,
grid = 30,
control = grid_ctrl
)
grid_results
While researching the error I came across this post but I don't believe I've enabled logging while using parallel processing
In the image below, notice the try-errr message:
Any help you can provide would be greatly appreciated.