I'm working with a dataset in R and I want to create a function that separates values into three categories based on quartiles. Values below the first quartile (Q1) should be labeled "Low expression", values above the third quartile (Q3) should be labeled "High expression", and everything else should be labeled "NA".
The purpose of that function is to create a dataset containing extreme expression values of a gene var_1 (quartiles Q1 and Q4) that I will correlate with several patient outcomes (variables 1:7). In order words, are extreme values of var_1 (Q1 and Q4) influencing the response of the variables 1 to 7?
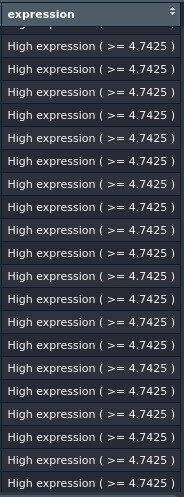
The problem is that the function keeps considering all observations of var_1 as high_expression. Can someone help me with this one?
function
fun_1 <- function(df, var_1) {
quants <- quantile(df[[var_1]], c(.25, .75))
tmp_1 <- df %>%
# selecting patients outcomes and gene expression
select(c(1:7) | {{var_1}}) %>%
# creating a new variable classifying Q1 and Q4
mutate(
expression =
case_when(
{{var_1}} >= quants[["75%"]] ~ sprintf(
"High expression ( >= %.02f )", quants[["75%"]]),
{{var_1}} <= quants[["25%"]] ~ sprintf(
"Low expression ( <= %.02f )", quants[["25%"]]),
.default = NA)) %>%
# excluding Q2 and Q3
filter(!is.na(expression))
tmp_1
}
fun_1(dataset_name, "gene_name")
output