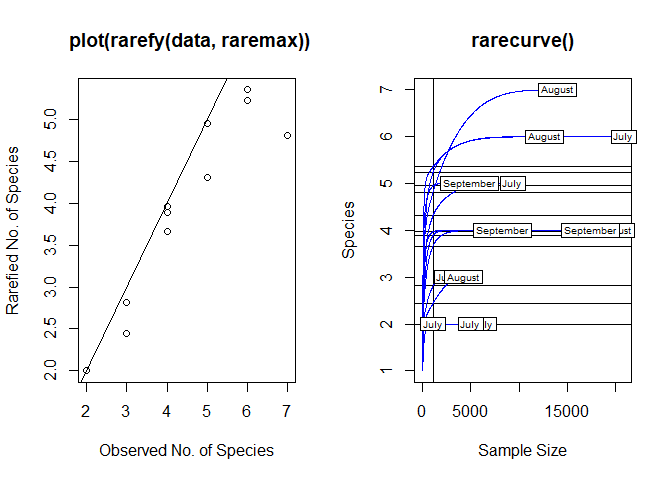
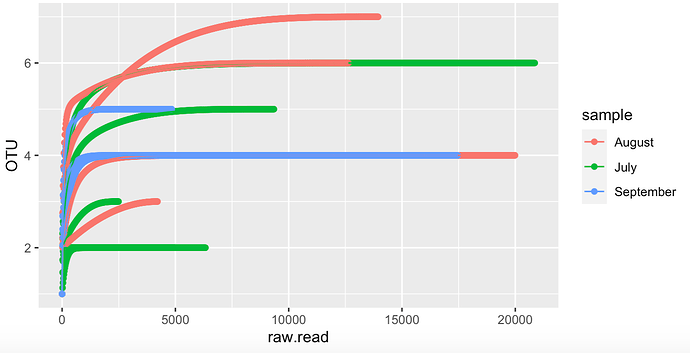
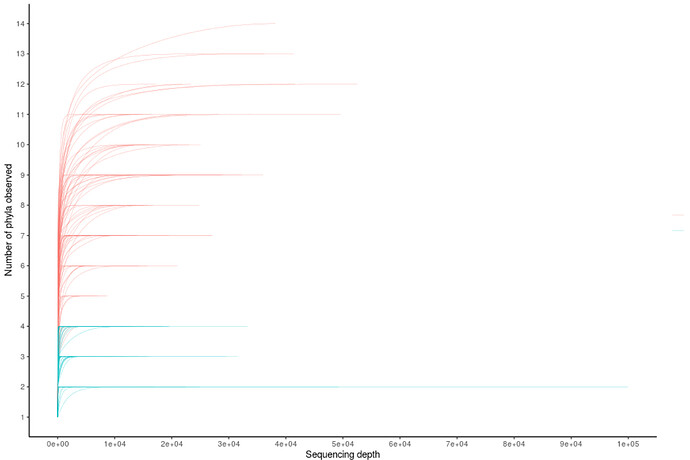
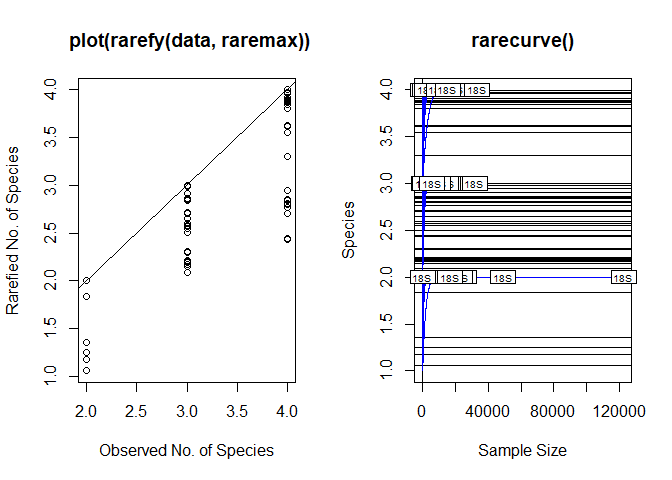
Thank you very much, using that function it worked, and now the graph looks better, although I am still not very convinced by the lines, I feel that they still look like weaves between them.
One last question JossChavez. , it is possible to establish the number of the axis of the Y, cause in my whole data (here its just a pice of it), I have approximately 29 OTU (the number of species that appear on the axis of the y) and when I obtain the graph shows up to 20 OTU, I would like the actual number of OTU to appear on the Y axis. Is it possible to set the axis number?. Sorry I'm very new in R and programming.
Thanks in advance
library(phyloseq)
library(tidyverse)
library(vegan)
#> Loading required package: permute
#> Loading required package: lattice
#> This is vegan 2.5-6
library(readxl)
clase <- tibble::tribble(
~OTU, ~`18S`, ~`18S`, ~`18S`, ~`18S`, ~`18S`, ~`18S`, ~`18S`, ~`18S`, ~`18S`, ~`18S`, ~`18S`, ~`18S`, ~`18S`, ~`18S`, ~`18S`, ~`18S`, ~`18S`, ~`18S`, ~`18S`, ~`18S`, ~`18S`, ~`18S`, ~`18S`, ~`18S`, ~`18S`, ~`18S`, ~`18S`, ~`18S`, ~`18S`, ~`18S`, ~`18S`, ~`18S`, ~`18S`, ~`18S`, ~`18S`, ~`18S`, ~`18S`, ~`18S`, ~`18S`, ~`18S`, ~`18S`, ~`18S`, ~`18S`, ~`18S`, ~`18S`, ~`18S`, ~`18S`, ~`18S`, ~`18S`, ~`18S`, ~`18S`, ~`18S`, ~`18S`, ~`18S`, ~`18S`, ~`18S`, ~`18S`, ~`18S`, ~`18S`, ~`18S`, ~`18S`, ~`18S`, ~`18S`, ~`18S`, ~`18S`, ~`18S`, ~`18S`, ~`18S`, ~`18S`, ~`18S`, ~`18S`, ~`18S`, ~`18S`, ~`18S`, ~`18S`, ~`18S`, ~`18S`, ~`18S`, ~`18S`, ~`18S`, ~`18S`, ~`18S`, ~`18S`, ~`18S`, ~`18S`, ~`18S`, ~`18S`, ~`18S`, ~`18S`, ~`18S`, ~`18S`, ~`18S`, ~`18S`,
"Alveolata", 0L, 0L, 0L, 36L, 0L, 6455L, 78L, 16L, 0L, 0L, 0L, 0L, 75L, 0L, 0L, 36L, 118L, 3799L, 0L, 0L, 297L, 230L, 198L, 761L, 0L, 227L, 0L, 510L, 40L, 0L, 0L, 0L, 0L, 20L, 0L, 30L, 363L, 45L, 2382L, 0L, 0L, 0L, 0L, 2069L, 0L, 0L, 0L, 0L, 0L, 0L, 74L, 140L, 186L, 0L, 183L, 277L, 0L, 99L, 648L, 2744L, 747L, 67L, 0L, 0L, 31L, 0L, 0L, 0L, 1099L, 0L, 41L, 0L, 0L, 113L, 469L, 37L, 0L, 0L, 0L, 0L, 111L, 816L, 0L, 0L, 342L, 0L, 0L, 0L, 874L, 0L, 107L, 531L, 0L,
"Amoebozoa", 0L, 0L, 44L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 52L, 0L, 3L, 0L, 0L, 0L, 0L, 0L, 4L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 16L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 13L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 673L, 0L, 130L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 38L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 3953L,
"Rhizaria", 0L, 0L, 0L, 44L, 0L, 0L, 0L, 66L, 0L, 0L, 0L, 0L, 0L, 8L, 0L, 31L, 0L, 119L, 0L, 0L, 12L, 0L, 0L, 77L, 26L, 105L, 0L, 0L, 0L, 53L, 0L, 0L, 0L, 0L, 12L, 7L, 150L, 12L, 0L, 0L, 0L, 10L, 0L, 0L, 67L, 68L, 0L, 73L, 0L, 0L, 56L, 0L, 30L, 7L, 34L, 6L, 0L, 0L, 0L, 0L, 654L, 39L, 0L, 0L, 0L, 0L, 96L, 24L, 677L, 0L, 0L, 0L, 0L, 0L, 0L, 5L, 0L, 12L, 0L, 0L, 0L, 0L, 0L, 20L, 0L, 8L, 0L, 19L, 312L, 0L, 0L, 389L, 0L,
"Stramenopiles", 35L, 1414L, 5272L, 2379L, 106L, 2066L, 393L, 7429L, 208L, 4266L, 25094L, 560L, 655L, 874L, 3673L, 267L, 699L, 482L, 1935L, 2221L, 84L, 297L, 401L, 1264L, 654L, 1370L, 439L, 305L, 1502L, 727L, 49170L, 223L, 2674L, 1436L, 449L, 684L, 867L, 511L, 5752L, 953L, 1053L, 83L, 710L, 385L, 1370L, 5062L, 122478L, 647L, 49L, 577L, 1176L, 1833L, 127L, 247L, 18175L, 7793L, 1270L, 9834L, 425L, 2067L, 4058L, 855L, 20948L, 1980L, 1991L, 1023L, 1342L, 1757L, 4962L, 1306L, 192L, 96L, 432L, 420L, 1914L, 316L, 312L, 9520L, 462L, 22327L, 1038L, 298L, 1702L, 157L, 844L, 319L, 106L, 7008L, 1002L, 13285L, 390L, 1724L, 705L,
"Un Eukaryota", 2150L, 862L, 395L, 201L, 617L, 7707L, 631L, 1955L, 2196L, 2616L, 4347L, 830L, 257L, 4696L, 2135L, 6700L, 5863L, 1686L, 2718L, 36L, 361L, 656L, 591L, 3106L, 553L, 2973L, 146L, 787L, 10534L, 2460L, 86L, 166L, 7L, 9024L, 3332L, 7256L, 6716L, 800L, 2268L, 744L, 723L, 247L, 3242L, 1103L, 7087L, 26475L, 68L, 82L, 117L, 605L, 11458L, 1959L, 1265L, 760L, 1256L, 10297L, 1552L, 5664L, 1050L, 2236L, 13820L, 10785L, 3914L, 3169L, 810L, 677L, 10142L, 12313L, 6801L, 205L, 898L, 95L, 313L, 362L, 4736L, 2484L, 3719L, 327L, 394L, 91L, 1078L, 1073L, 883L, 1943L, 631L, 187L, 3L, 2244L, 12976L, 3626L, 1062L, 30639L, 1086L
)
data <- clase
data # data is the name of your matrix
#> # A tibble: 5 x 94
#> OTU `18S` `18S` `18S` `18S` `18S` `18S` `18S` `18S` `18S` `18S` `18S` `18S`
#> <chr> <int> <int> <int> <int> <int> <int> <int> <int> <int> <int> <int> <int>
#> 1 Alveo~ 0 0 0 36 0 6455 78 16 0 0 0 0
#> 2 Amoeb~ 0 0 44 0 0 0 0 0 0 0 52 0
#> 3 Rhiza~ 0 0 0 44 0 0 0 66 0 0 0 0
#> 4 Stram~ 35 1414 5272 2379 106 2066 393 7429 208 4266 25094 560
#> 5 Un Eu~ 2150 862 395 201 617 7707 631 1955 2196 2616 4347 830
#> # ... with 81 more variables: `18S` <int>, `18S` <int>, `18S` <int>,
#> # `18S` <int>, `18S` <int>, `18S` <int>, `18S` <int>, `18S` <int>,
#> # `18S` <int>, `18S` <int>, `18S` <int>, `18S` <int>, `18S` <int>,
#> # `18S` <int>, `18S` <int>, `18S` <int>, `18S` <int>, `18S` <int>,
#> # `18S` <int>, `18S` <int>, `18S` <int>, `18S` <int>, `18S` <int>,
#> # `18S` <int>, `18S` <int>, `18S` <int>, `18S` <int>, `18S` <int>,
#> # `18S` <int>, `18S` <int>, `18S` <int>, `18S` <int>, `18S` <int>,
#> # `18S` <int>, `18S` <int>, `18S` <int>, `18S` <int>, `18S` <int>,
#> # `18S` <int>, `18S` <int>, `18S` <int>, `18S` <int>, `18S` <int>,
#> # `18S` <int>, `18S` <int>, `18S` <int>, `18S` <int>, `18S` <int>,
#> # `18S` <int>, `18S` <int>, `18S` <int>, `18S` <int>, `18S` <int>,
#> # `18S` <int>, `18S` <int>, `18S` <int>, `18S` <int>, `18S` <int>,
#> # `18S` <int>, `18S` <int>, `18S` <int>, `18S` <int>, `18S` <int>,
#> # `18S` <int>, `18S` <int>, `18S` <int>, `18S` <int>, `18S` <int>,
#> # `18S` <int>, `18S` <int>, `18S` <int>, `18S` <int>, `18S` <int>,
#> # `18S` <int>, `18S` <int>, `18S` <int>, `18S` <int>, `18S` <int>,
#> # `18S` <int>, `18S` <int>, `18S` <int>
attach(clase)
rwnames <- OTU
data <- data.matrix(data[,-1])
rownames(data) <- rwnames
data <- t(data)
S <- specnumber(data)
raremax <- min(rowSums(data))
Srare <- rarefy(data, raremax)
#Plot rarefaction results
par(mfrow = c(1,2))
plot(S, Srare, xlab = "Observed No. of Species",
ylab = "Rarefied No. of Species",
main = " plot(rarefy(data, raremax))")
abline(0, 1)
rarecurve(data, step = 20,
sample = raremax,
col = "blue",
cex = 0.6,
main = "rarecurve()")
rarecurve(data[1:93,], step = 20, sample = raremax, col = "blue", cex = 0.6,
main = "rarecurve() on subset of data")
out <- rarecurve(data, step = 20, sample = raremax, label = T)
#Clean the list up a bit:
rare <- lapply(out, function(x){b <- as.data.frame(x)
b <- data.frame(clase = b[,1], raw.read = rownames(b))
b$raw.read <- as.numeric(gsub("N", "", b$raw.read))
return(b)})
#label list
names(rare) <- rownames(data)
#convert to data frame:
rare <- map_dfr(rare, function(x){
z <- data.frame(x)
return(z)
}, .id = "Sample")
write.csv(rare, file = "~/RSTUDIO/Bacteria-total/bacteria-clase-total-otu-rarefaction.csv")
#agregar manual el tratamiento
rarefaction <- read.csv("~/RSTUDIO/Bacteria-total/bacteria-clase-total-otu-rarefaction.csv", row.names=1)
df1 <- rarefaction
#ALTO
library(phyloseq)
library(tidyverse)
library(vegan)
library(readxl)
clase <- tibble::tribble(
~OTU, ~`16S`, ~`16S`, ~`16S`, ~`16S`, ~`16S`, ~`16S`, ~`16S`, ~`16S`, ~`16S`, ~`16S`, ~`16S`, ~`16S`, ~`16S`, ~`16S`, ~`16S`, ~`16S`, ~`16S`, ~`16S`, ~`16S`, ~`16S`, ~`16S`, ~`16S`, ~`16S`, ~`16S`, ~`16S`, ~`16S`, ~`16S`, ~`16S`, ~`16S`, ~`16S`, ~`16S`, ~`16S`, ~`16S`, ~`16S`, ~`16S`, ~`16S`, ~`16S`, ~`16S`, ~`16S`, ~`16S`, ~`16S`, ~`16S`, ~`16S`, ~`16S`, ~`16S`, ~`16S`, ~`16S`, ~`16S`, ~`16S`, ~`16S`, ~`16S`, ~`16S`, ~`16S`, ~`16S`, ~`16S`, ~`16S`, ~`16S`, ~`16S`, ~`16S`, ~`16S`, ~`16S`, ~`16S`, ~`16S`, ~`16S`, ~`16S`, ~`16S`, ~`16S`, ~`16S`, ~`16S`, ~`16S`, ~`16S`, ~`16S`, ~`16S`, ~`16S`, ~`16S`, ~`16S`, ~`16S`, ~`16S`, ~`16S`, ~`16S`, ~`16S`, ~`16S`, ~`16S`, ~`16S`, ~`16S`, ~`16S`, ~`16S`, ~`16S`, ~`16S`, ~`16S`, ~`16S`, ~`16S`, ~`16S`, ~`16S`, ~`16S`, ~`16S`,
"Acidobacteria", 0L, 0L, 0L, 0L, 0L, 9L, 0L, 4L, 0L, 0L, 0L, 0L, 0L, 4L, 0L, 19L, 0L, 50L, 0L, 0L, 0L, 0L, 0L, 0L, 8L, 0L, 0L, 0L, 0L, 0L, 0L, 36L, 0L, 0L, 0L, 10L, 0L, 11L, 27L, 0L, 9L, 0L, 0L, 0L, 0L, 0L, 0L, 55L, 0L, 0L, 0L, 0L, 2L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 8L, 0L, 50L, 0L, 0L, 0L, 0L, 5L, 2L, 34L, 19L, 164L, 3L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 2L, 0L, 0L, 6L, 0L, 0L, 0L, 0L, 32L, 0L, 0L, 82L, 63L,
"Actinobacteria", 389L, 33L, 31L, 797L, 3864L, 9570L, 878L, 338L, 4450L, 1819L, 2093L, 1163L, 1988L, 719L, 2646L, 4894L, 771L, 90L, 42L, 533L, 2440L, 1357L, 3194L, 12L, 10548L, 2370L, 669L, 3716L, 4786L, 82L, 1482L, 5804L, 4329L, 34L, 0L, 2174L, 93L, 2141L, 3592L, 1346L, 7088L, 4521L, 808L, 338L, 4087L, 6805L, 1020L, 3670L, 2199L, 95L, 54L, 427L, 2870L, 331L, 3372L, 636L, 3906L, 8550L, 277L, 9361L, 4837L, 6891L, 8241L, 3089L, 90L, 62L, 659L, 13399L, 669L, 2628L, 1421L, 2630L, 2003L, 1218L, 668L, 107L, 5767L, 2243L, 615L, 239L, 17L, 157L, 216L, 6913L, 3364L, 336L, 437L, 109L, 1321L, 1131L, 980L, 7694L, 179L, 7413L, 9547L, 4713L,
"Armatimonadetes", 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 4L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L,
"BRC1", 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 20L, 0L, 4L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 21L, 0L, 0L, 0L, 0L, 0L, 0L, 7L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 8L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 3L, 15L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 4L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 2L, 0L,
"Bacteroidetes", 5936L, 4927L, 1035L, 1690L, 5390L, 10966L, 3329L, 12756L, 15453L, 10255L, 15262L, 3485L, 6123L, 9240L, 11010L, 14675L, 6400L, 298L, 3043L, 2099L, 3060L, 4345L, 4240L, 525L, 15187L, 13848L, 5335L, 6041L, 5490L, 508L, 9433L, 28803L, 6488L, 3842L, 302L, 8097L, 9782L, 13500L, 1058L, 8161L, 16490L, 19264L, 4630L, 7699L, 3957L, 10869L, 6708L, 40145L, 5246L, 1954L, 3696L, 23605L, 15779L, 14385L, 142L, 1906L, 15517L, 22258L, 10028L, 7378L, 4426L, 17895L, 1204L, 14137L, 3145L, 8091L, 1632L, 9577L, 20608L, 45241L, 2551L, 3660L, 11260L, 14090L, 2150L, 10691L, 6029L, 11502L, 2311L, 2757L, 86L, 781L, 3277L, 5658L, 8732L, 4174L, 1650L, 13487L, 28515L, 11051L, 17648L, 1852L, 6891L, 11030L, 19868L, 29711L,
"Saccharibacteria", 0L, 0L, 0L, 0L, 4L, 0L, 0L, 3L, 0L, 5L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 6L, 0L, 0L, 0L, 0L, 0L, 11L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 4L, 0L, 0L, 0L, 0L, 6L, 0L, 2L, 0L, 0L, 3L, 0L, 0L, 0L, 0L, 0L, 0L, 4L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 5L, 0L, 0L, 0L, 0L, 0L, 7L, 10L,
"Chlamydiae", 0L, 0L, 0L, 3L, 67L, 148L, 0L, 5L, 61L, 208L, 67L, 60L, 0L, 3L, 36L, 108L, 0L, 6L, 0L, 0L, 24L, 14L, 22L, 0L, 298L, 254L, 26L, 84L, 30L, 0L, 15L, 126L, 0L, 0L, 0L, 0L, 0L, 29L, 20L, 0L, 168L, 122L, 76L, 0L, 22L, 156L, 15L, 40L, 29L, 0L, 0L, 0L, 22L, 4L, 4L, 0L, 54L, 283L, 3L, 122L, 52L, 252L, 93L, 4L, 0L, 2L, 0L, 56L, 5L, 112L, 31L, 9L, 210L, 104L, 39L, 0L, 64L, 10L, 4L, 0L, 0L, 3L, 9L, 17L, 101L, 0L, 7L, 0L, 75L, 38L, 59L, 94L, 0L, 32L, 146L, 69L,
"Chloroflexi", 0L, 0L, 0L, 0L, 22L, 129L, 0L, 76L, 0L, 243L, 37L, 119L, 25L, 35L, 124L, 118L, 281L, 359L, 0L, 0L, 0L, 35L, 0L, 0L, 440L, 87L, 0L, 0L, 0L, 0L, 18L, 223L, 56L, 0L, 0L, 2L, 0L, 44L, 74L, 0L, 93L, 90L, 44L, 0L, 18L, 52L, 32L, 250L, 0L, 6L, 0L, 314L, 108L, 160L, 0L, 6L, 81L, 440L, 24L, 96L, 253L, 338L, 310L, 67L, 0L, 56L, 2L, 241L, 125L, 393L, 47L, 1222L, 202L, 68L, 0L, 65L, 45L, 2L, 28L, 0L, 0L, 0L, 0L, 74L, 0L, 10L, 7L, 4L, 135L, 111L, 0L, 228L, 0L, 35L, 633L, 289L,
"Cloacimonetes", 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 3L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 2L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 12L, 17L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L,
"Cyanobacteria", 0L, 0L, 0L, 0L, 0L, 37L, 2L, 761L, 22L, 102L, 0L, 12L, 163L, 3L, 81L, 111L, 131L, 645L, 0L, 0L, 0L, 0L, 0L, 0L, 52L, 63L, 0L, 0L, 8L, 0L, 69L, 153L, 25L, 0L, 0L, 27L, 17L, 55L, 92L, 26L, 66L, 0L, 0L, 0L, 0L, 4L, 67L, 388L, 349L, 0L, 0L, 128L, 63L, 285L, 34L, 8L, 16L, 0L, 31L, 12L, 145L, 97L, 798L, 137L, 0L, 2L, 0L, 24L, 266L, 150L, 77L, 5931L, 66L, 14L, 0L, 47L, 62L, 4L, 51L, 0L, 0L, 0L, 177L, 3L, 20L, 0L, 2L, 0L, 28L, 0L, 0L, 360L, 0L, 18L, 509L, 645L,
"Elusimicrobia", 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 2L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L,
"Firmicutes", 39L, 371L, 467L, 918L, 550L, 79L, 182L, 1582L, 3115L, 3272L, 1058L, 2009L, 551L, 357L, 402L, 656L, 792L, 121L, 0L, 805L, 3L, 2L, 0L, 0L, 4229L, 2930L, 403L, 998L, 63L, 131L, 1780L, 4215L, 2657L, 207L, 222L, 23L, 21L, 265L, 628L, 5899L, 3364L, 1670L, 710L, 720L, 34L, 53L, 5536L, 3207L, 2263L, 287L, 465L, 2224L, 1899L, 241L, 1198L, 2157L, 1789L, 736L, 315L, 3956L, 689L, 74L, 3726L, 1788L, 451L, 308L, 1401L, 3413L, 528L, 2398L, 2548L, 73L, 1430L, 539L, 1118L, 2410L, 25L, 449L, 101L, 557L, 2796L, 106L, 2018L, 1568L, 2L, 124L, 163L, 616L, 1295L, 862L, 1550L, 1752L, 63L, 46L, 3455L, 4248L,
"Fusobacteria", 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 2L, 0L,
"Gemmatimonadetes", 0L, 0L, 0L, 0L, 0L, 80L, 0L, 0L, 12L, 4L, 0L, 10L, 0L, 0L, 3L, 19L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 15L, 0L, 0L, 13L, 0L, 0L, 0L, 36L, 0L, 0L, 0L, 0L, 0L, 9L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 48L, 3L, 8L, 0L, 0L, 0L, 0L, 13L, 0L, 0L, 0L, 6L, 11L, 0L, 0L, 4L, 33L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 23L, 20L, 2L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 9L, 0L, 0L, 0L, 0L, 0L, 0L, 19L, 0L, 20L, 36L, 14L,
"Hydrogenedentes", 0L, 0L, 0L, 0L, 7L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 4L, 10L, 19L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 250L, 0L, 0L, 0L, 0L, 0L, 9L, 24L, 0L, 0L, 0L, 0L, 0L, 11L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 10L, 9L, 6L, 0L, 0L, 0L, 0L, 19L, 0L, 0L, 0L, 0L, 0L, 0L, 32L, 0L, 0L, 12L, 9L, 0L, 0L, 0L, 5L, 68L, 23L, 18L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 5L, 0L, 0L, 0L, 0L, 16L, 0L, 0L, 29L, 3L,
"Verrucomicrobia", 108L, 0L, 50L, 0L, 949L, 2781L, 318L, 397L, 953L, 651L, 811L, 566L, 117L, 304L, 523L, 1272L, 469L, 42L, 0L, 0L, 349L, 456L, 791L, 0L, 3072L, 833L, 222L, 1019L, 743L, 0L, 350L, 1170L, 5L, 28L, 25L, 471L, 48L, 796L, 562L, 95L, 1027L, 1162L, 391L, 58L, 517L, 2086L, 266L, 1583L, 85L, 0L, 0L, 216L, 757L, 222L, 295L, 0L, 974L, 2708L, 111L, 1706L, 1167L, 2037L, 1319L, 686L, 0L, 111L, 0L, 714L, 654L, 1105L, 549L, 111L, 566L, 543L, 311L, 112L, 1421L, 576L, 105L, 100L, 4L, 29L, 52L, 658L, 1017L, 2L, 171L, 34L, 474L, 551L, 541L, 1398L, 51L, 1307L, 2395L, 885L,
"Un Bacteria", 24L, 54L, 11L, 0L, 334L, 1223L, 60L, 127L, 718L, 496L, 130L, 193L, 76L, 72L, 110L, 793L, 121L, 174L, 0L, 0L, 175L, 200L, 181L, 0L, 2145L, 410L, 42L, 139L, 478L, 0L, 114L, 751L, 239L, 4L, 0L, 197L, 66L, 480L, 222L, 172L, 685L, 188L, 106L, 9L, 180L, 962L, 180L, 261L, 158L, 0L, 65L, 196L, 327L, 123L, 55L, 23L, 690L, 1022L, 62L, 685L, 749L, 1024L, 715L, 254L, 10L, 21L, 0L, 805L, 384L, 431L, 216L, 632L, 355L, 146L, 56L, 68L, 295L, 283L, 128L, 18L, 0L, 4L, 18L, 455L, 432L, 7L, 27L, 16L, 410L, 343L, 193L, 774L, 36L, 309L, 1392L, 958L
)
data <- clase
data # data is the name of your matrix
#> # A tibble: 17 x 97
#> OTU `16S` `16S` `16S` `16S` `16S` `16S` `16S` `16S` `16S` `16S` `16S` `16S`
#> <chr> <int> <int> <int> <int> <int> <int> <int> <int> <int> <int> <int> <int>
#> 1 Acid~ 0 0 0 0 0 9 0 4 0 0 0 0
#> 2 Acti~ 389 33 31 797 3864 9570 878 338 4450 1819 2093 1163
#> 3 Arma~ 0 0 0 0 0 0 0 0 0 0 0 0
#> 4 BRC1 0 0 0 0 0 0 0 0 0 0 0 0
#> 5 Bact~ 5936 4927 1035 1690 5390 10966 3329 12756 15453 10255 15262 3485
#> 6 Sacc~ 0 0 0 0 4 0 0 3 0 5 0 0
#> 7 Chla~ 0 0 0 3 67 148 0 5 61 208 67 60
#> 8 Chlo~ 0 0 0 0 22 129 0 76 0 243 37 119
#> 9 Cloa~ 0 0 0 0 0 0 0 0 0 0 0 0
#> 10 Cyan~ 0 0 0 0 0 37 2 761 22 102 0 12
#> 11 Elus~ 0 0 0 0 0 0 0 0 0 0 0 0
#> 12 Firm~ 39 371 467 918 550 79 182 1582 3115 3272 1058 2009
#> 13 Fuso~ 0 0 0 0 0 0 0 0 0 0 0 0
#> 14 Gemm~ 0 0 0 0 0 80 0 0 12 4 0 10
#> 15 Hydr~ 0 0 0 0 7 0 0 0 0 0 0 0
#> 16 Verr~ 108 0 50 0 949 2781 318 397 953 651 811 566
#> 17 Un B~ 24 54 11 0 334 1223 60 127 718 496 130 193
#> # ... with 84 more variables: `16S` <int>, `16S` <int>, `16S` <int>,
#> # `16S` <int>, `16S` <int>, `16S` <int>, `16S` <int>, `16S` <int>,
#> # `16S` <int>, `16S` <int>, `16S` <int>, `16S` <int>, `16S` <int>,
#> # `16S` <int>, `16S` <int>, `16S` <int>, `16S` <int>, `16S` <int>,
#> # `16S` <int>, `16S` <int>, `16S` <int>, `16S` <int>, `16S` <int>,
#> # `16S` <int>, `16S` <int>, `16S` <int>, `16S` <int>, `16S` <int>,
#> # `16S` <int>, `16S` <int>, `16S` <int>, `16S` <int>, `16S` <int>,
#> # `16S` <int>, `16S` <int>, `16S` <int>, `16S` <int>, `16S` <int>,
#> # `16S` <int>, `16S` <int>, `16S` <int>, `16S` <int>, `16S` <int>,
#> # `16S` <int>, `16S` <int>, `16S` <int>, `16S` <int>, `16S` <int>,
#> # `16S` <int>, `16S` <int>, `16S` <int>, `16S` <int>, `16S` <int>,
#> # `16S` <int>, `16S` <int>, `16S` <int>, `16S` <int>, `16S` <int>,
#> # `16S` <int>, `16S` <int>, `16S` <int>, `16S` <int>, `16S` <int>,
#> # `16S` <int>, `16S` <int>, `16S` <int>, `16S` <int>, `16S` <int>,
#> # `16S` <int>, `16S` <int>, `16S` <int>, `16S` <int>, `16S` <int>,
#> # `16S` <int>, `16S` <int>, `16S` <int>, `16S` <int>, `16S` <int>,
#> # `16S` <int>, `16S` <int>, `16S` <int>, `16S` <int>, `16S` <int>,
#> # `16S` <int>
attach(clase)
#> The following object is masked from clase (pos = 3):
#>
#> OTU
rwnames <- OTU
data <- data.matrix(data[,-1])
rownames(data) <- rwnames
data <- t(data)
S <- specnumber(data)
raremax <- min(rowSums(data))
Srare <- rarefy(data, raremax)
#Plot rarefaction results
par(mfrow = c(1,2))
plot(S, Srare, xlab = "Observed No. of Species",
ylab = "Rarefied No. of Species",
main = " plot(rarefy(data, raremax))")
abline(0, 1)
rarecurve(data, step = 20,
sample = raremax,
col = "blue",
cex = 0.6,
main = "rarecurve()")

rarecurve(data[1:96,], step = 20, sample = raremax, col = "blue", cex = 0.6,
main = "rarecurve() on subset of data")
out <- rarecurve(data, step = 20, sample = raremax, label = T)

#Clean the list up a bit:
rare <- lapply(out, function(x){b <- as.data.frame(x)
b <- data.frame(clase = b[,1], raw.read = rownames(b))
b$raw.read <- as.numeric(gsub("N", "", b$raw.read))
return(b)})
#label list
names(rare) <- rownames(data)
#convert to data frame:
rare <- map_dfr(rare, function(x){
z <- data.frame(x)
return(z)
}, .id = "Sample")
write.csv(rare, file = "~/RSTUDIO/Bacteria-total/bacteria-clase-total-otu-rarefactioNn.csv")
#agregar manual el tratamiento
rarefactioNn <- read.csv("~/RSTUDIO/Bacteria-total/bacteria-clase-total-otu-rarefactioNn.csv", row.names=1)
df2 <- rarefactioNn
df=rbind(df1,df2)
df$group = rep(1, nrow(df) )
n = 0
for (i in 1:nrow(df) ) {
if (df$raw.read[i] == 1) {
n = n + 1
}
df$group[i] <- n
}
library(ggplot2)
#plot
#generar una escala de colores
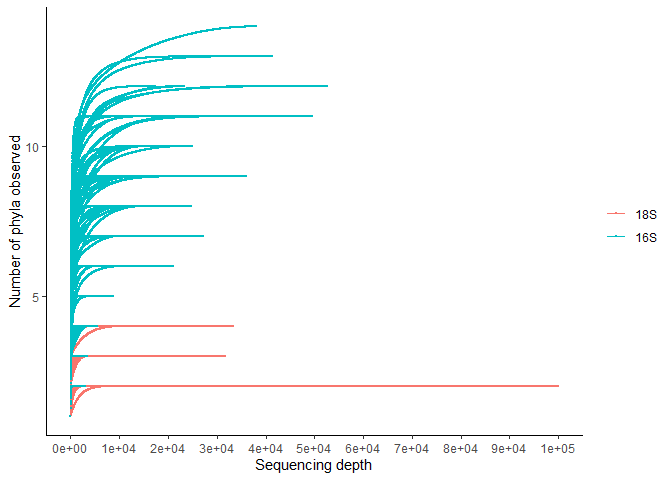
rarcu <- ggplot(data=df, aes(x=raw.read, y=clase, color = factor (Sample), group = group ))+
geom_line(size=0.1)+
geom_point(size=0.1)
rarcu + theme_classic()+
theme(legend.title = element_blank())+
scale_x_continuous(labels = scales::scientific_format())+
ylab("Number of phyla observed")+
xlab("Sequencing depth")+
scale_x_continuous(limits = c(0, 100000), breaks = c(0, 10000, 20000, 30000, 40000, 50000, 60000, 70000, 80000, 90000, 100000))
#> Scale for 'x' is already present. Adding another scale for 'x', which will
#> replace the existing scale.
#> Warning: Removed 1129 row(s) containing missing values (geom_path).
#> Warning: Removed 1129 rows containing missing values (geom_point).
Created on 2020-05-16 by the reprex package (v0.3.0)