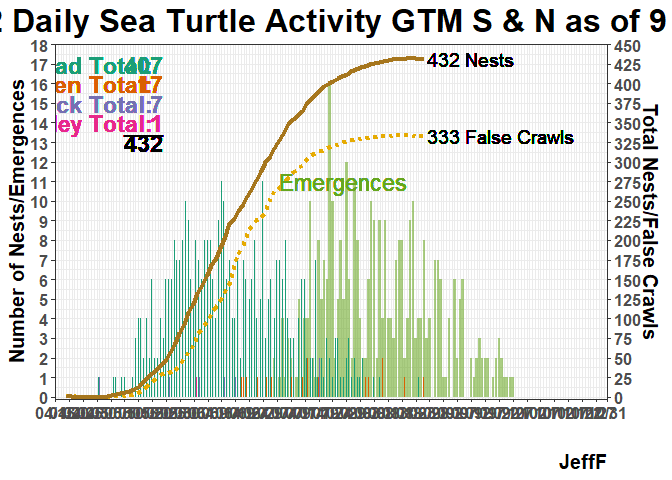
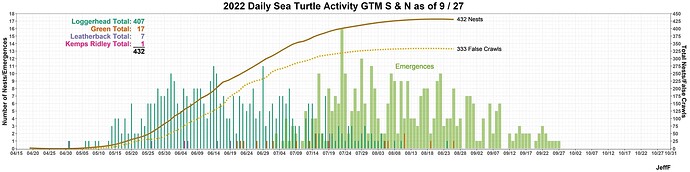
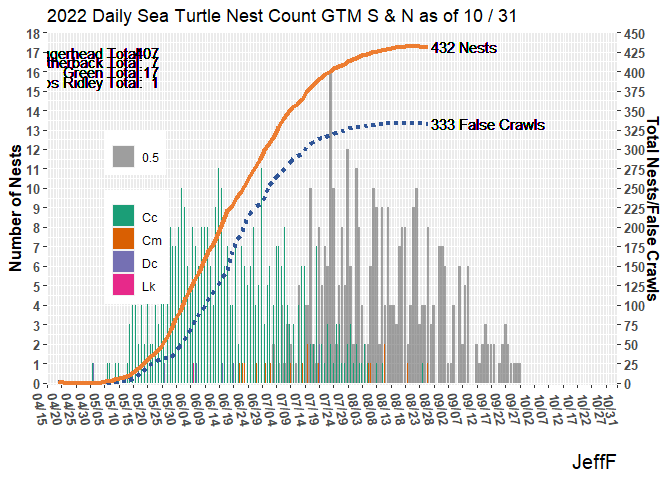
I’ve been hunting around for a way to do this, if even possible. In the plot below is there a way the geom_text annotations such as Loggerhead Total: 407 matches the color for “Cc” which is part of “species” used to specify fill color for the bar plots..
Thanks,
Jeff
library(tidyverse)
library("lubridate")
#> Loading required package: timechange
#>
#> Attaching package: 'lubridate'
#> The following objects are masked from 'package:base':
#>
#> date, intersect, setdiff, union
library("RColorBrewer")
library("reprex")
start_date <- ymd("2022-04-15")
end_date <- ymd("2022-10-31")
position_date <- ymd("2022-05-05") #place on x-axis
last_report_date <- ymd("2022-10-31")
date_length <-days(start_date %--% end_date )
position_date_len <- days(start_date %--% position_date)
relative_date <- position_date_len/date_length
rel_y <- 13/18
coeff <- 25
turtle_activity_gtm <- read_csv("https://www.dropbox.com/s/7ubvhajvkhx53kc/mpt_act_rep_cum_fc_cum_nest.csv?dl=1")
#> Rows: 765 Columns: 66
#> ── Column specification ────────────────────────────────────────────────────────
#> Delimiter: ","
#> chr (22): beach, county, activity, ref_no, activity_comments, encountered?,...
#> dbl (35): uid, activity_no, fcrawl, fcrawl_cum, clutch, clutch_cum, nest_no...
#> lgl (6): final_treatment, light_management, relocation_reason, lost_nest, ...
#> date (3): activity_date, emerge_date, inventory_date
#>
#> ℹ Use `spec()` to retrieve the full column specification for this data.
#> ℹ Specify the column types or set `show_col_types = FALSE` to quiet this message.
#Above file has cumulative false crawls and cumulative nests added along with proper date format.
cc_total <- count(turtle_activity_gtm |> filter(activity=="N") |> filter(species =='Cc'))
dc_total <- count(turtle_activity_gtm |> filter(activity=="N") |> filter(species =='Dc'))
cm_total <- count(turtle_activity_gtm |> filter(activity=="N") |> filter(species =='Cm'))
lk_total <- count(turtle_activity_gtm |> filter(activity=="N") |> filter(species =='Lk'))
#suppressWarnings(print(
ggplot(data = (turtle_activity_gtm |> filter(activity=="N"))) +
scale_fill_brewer(palette = "Dark2") +
scale_color_brewer(palette = "Dark2") +
geom_bar(aes(x = emerge_date, alpha=.5),
na.rm = TRUE,
position = position_dodge2(preserve = "total")) +
geom_bar(aes(x = activity_date, fill = species),
na.rm = TRUE,
position = position_dodge2(preserve = "single")) +
scale_y_continuous(breaks = 0:18, expand = c(0,0),
sec.axis = sec_axis(~.*coeff, name="Total Nests/False Crawls",
breaks = seq(0,450,25))) +
expand_limits(y = c(-.1, 18)) +
scale_x_date(breaks = c(seq.Date((start_date), (end_date),
by = '5 days'), end_date),
date_labels = "%m/%d",
date_minor_breaks = "1 day",
limits = c( start_date, end_date),
expand = c(0,0)) +
labs(x="",y="Number of Nests", title=paste("2022 Daily Sea Turtle Nest Count GTM S & N as of",month(last_report_date),"/",mday(last_report_date)),
caption='JeffF') +
theme(axis.text.y = element_text(face = "bold",
size = 10, angle = 0),
axis.text.x = element_text(face = "bold",
size = 10, angle = -80),
legend.position=c(x=relative_date, y=rel_y),
legend.justification = c(0, 1), #upper left
legend.title = element_blank(),
axis.title = element_text(size = 12, face = "bold"),
plot.caption = element_text(size = 15)) +
#theme(legend.position = "none") +
geom_smooth(data = turtle_activity_gtm, method = 'loess', formula = 'y ~ x',
aes(x=activity_date, y=fcrawl_cum/coeff),linetype = "111111",
linewidth = 1.5, color = '#2F5597', span = 0.08, se = FALSE) +
geom_smooth(data = turtle_activity_gtm, method = 'loess', formula = 'y ~ x',
aes(x=activity_date, y=clutch_cum/coeff), linewidth = 1.5,
color ='#ED7D31', span = 0.08, se = FALSE) +
geom_text(x=ymd(max(turtle_activity_gtm$activity_date+1,na.rm = TRUE)),
y=18*(max(turtle_activity_gtm$fcrawl_cum/450)),
hjust = "left",label=paste(max(turtle_activity_gtm$fcrawl_cum),
'False Crawls' )) +
geom_text(x=ymd(max(turtle_activity_gtm$activity_date+1,na.rm = TRUE)),
y=18*(max(turtle_activity_gtm$clutch_cum/450)),
hjust = "left",label=paste(max(turtle_activity_gtm$clutch_cum),
'Nests')) +
geom_text(x=ymd("2022-05-20"), y=17, hjust = "right",
label=paste("Loggerhead Total: ")) +
geom_text(x=ymd("2022-05-20")+days(4), y=17, hjust = "right",
label=paste(cc_total)) +
geom_text(x=ymd("2022-05-20"), y=16.5, hjust = "right",
label=paste("Leatherback Total: ")) +
geom_text(x=ymd("2022-05-20")+days(4), y=16.5, hjust = "right",
label=paste(dc_total)) +
geom_text(x=ymd("2022-05-20"), y=16, hjust = "right",
label=paste("Green Total: ")) +
geom_text(x=ymd("2022-05-20")+days(4), y=16, hjust = "right",
label=paste(cm_total)) +
geom_text(x=ymd("2022-05-20"), y=15.5, hjust = "right",
label=paste("Kemps Ridley Total: ")) +
geom_text(x=ymd("2022-05-20")+days(4), y=15.5, hjust = "right",
label=paste(lk_total))
#> Warning: Removed 1 rows containing non-finite values (`stat_smooth()`).
#> Removed 1 rows containing non-finite values (`stat_smooth()`).
Created on 2023-01-18 with reprex v2.0.2