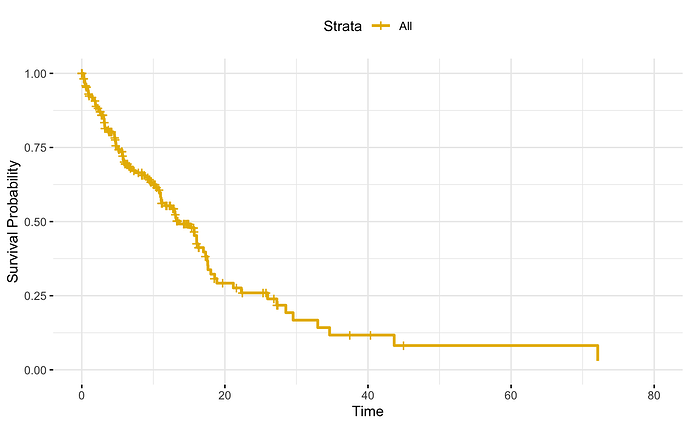
I'm working on some survival analysis using a cox model with a single variable. I would like to plot the survival curves of the two but am struggling.
library(tidyverse)
library(survival)
library(survminer)
set.seed(123)
pdata <- data.frame(
time = rexp(200, rate = 0.1), # Survival times
status = sample(0:1, 200, replace = TRUE), # Censoring indicator
sub_plan = factor(sample(c("pro", "standard"), 200, replace = TRUE)) # Subscription plans
)
fit.cox <- coxph(Surv(time, status) ~ sub_plan, data = pdata)
surv_estimates <- survfit(fit.cox)
ggsurvplot(
surv_estimates,
data = pdata,
pval = FALSE,
conf.int = FALSE,
palette = c("#E7B800", "#2E9FDF"),
xlab = "Time",
ylab = "Survival Probability",
ggtheme = theme_minimal()
)
This results in the following plot:
Hoped/expected/wouldlike two curves but just get a single one, one for sub_plan 'pro' and one for 'standard'. Can I do this using my fit and ggsurvplot?
dougfir:
ggsurvplot(
surv_estimates,
data = pdata,
pval = FALSE,
conf.int = FALSE,
palette = c("#E7B800", "#2E9FDF"),
xlab = "Time",
ylab = "Survival Probability",
ggtheme = theme_minimal()
)
What happens if you add color = sub_plan as one of the arguments to ggsurvplot()?
I was able to get this working with some help from a datacamp lesson:
fit.cox <- coxph(Surv(time, status) ~ sub_plan, data = pdata)
new_pdata <- pdata |> distinct(sub_plan)
surv_estimates <- survfit(fit.cox, data = pdata, newdata = new_pdata, conf.type = "none")
ggsurvplot(
surv_estimates,
data = pdata,
pval = FALSE,
conf.int = FALSE,
palette = c("#E7B800", "#2E9FDF"),
xlab = "Time",
ylab = "Survival Probability",
ggtheme = theme_minimal()
)
So surv_estimates has to be passed the fitted model, existing data pdata and also new_data with the distinct values of sub_plan
dromano:
color = sub_plan
I tried adding color = sub_plan but ggsurvplot complained about not recognizing sub_plan. Anyway, was able to get it working, see solution. Thanks all the same!
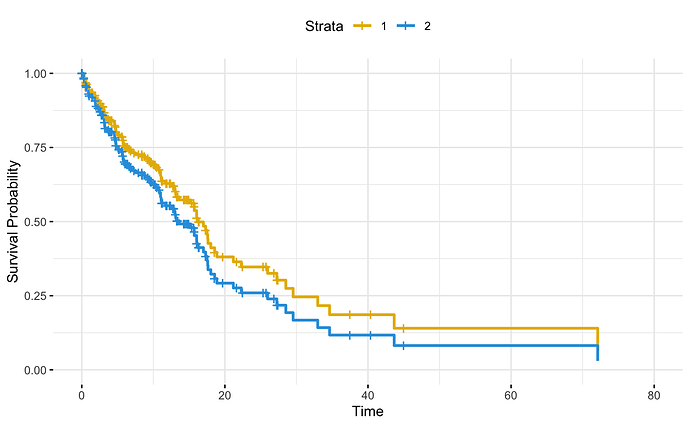
You shouldn't have to use newdata when you have a single table, and I would think those curves look suspiciously similar. Using the coxph() function results in a loss of the sub_plan identifier, but even so, the strata can be preserved through the strata() function, like this:
library(tidyverse)
library(survival)
library(survminer)
set.seed(123)
pdata <- data.frame(
time = rexp(200, rate = 0.1), # Survival times
status = sample(0:1, 200, replace = TRUE), # Censoring indicator
sub_plan = factor(sample(c("pro", "standard"), 200, replace = TRUE)) # Subscription plans
)
fit.cox <- coxph(Surv(time, status) ~ strata(sub_plan), data = pdata)
surv_estimates <- survfit(fit.cox)
ggsurvplot(
surv_estimates,
data = pdata,
pval = FALSE,
conf.int = FALSE,
palette = c("#E7B800", "#2E9FDF"),
xlab = "Time",
ylab = "Survival Probability",
ggtheme = theme_minimal()
)
Created on 2024-05-11 with reprex v2.0.2
1 Like
Interesting. Thanks for this. With your model:
fit.cox |> summary()
Call: coxph(formula = Surv(time, status) ~ strata(sub_plan), data = pdata)
Null model
log likelihood= -349.3911
n= 200
With the model not using strata():
fit.cox |> summary()
Call:
coxph(formula = Surv(time, status) ~ sub_plan, data = pdata)
n= 200, number of events= 96
coef exp(coef) se(coef) z Pr(>|z|)
sub_planstandard -0.2423 0.7848 0.2059 -1.177 0.239
exp(coef) exp(-coef) lower .95 upper .95
sub_planstandard 0.7848 1.274 0.5242 1.175
Concordance= 0.527 (se = 0.03 )
Likelihood ratio test= 1.39 on 1 df, p=0.2
Wald test = 1.39 on 1 df, p=0.2
Score (logrank) test = 1.39 on 1 df, p=0.2
It's a different model no? I do want the coefficients for sub_plan? Am I misunderstanding?
dougfir:
sub_planstandard
This is the effect of the coxph() function, which collapses the two subplans into a single one called "sub_planstandard" if you don't use strata() to treat the subplans as distinct. This is why you got a single curve in your original post. Without strata(), the distinction is lost.
1 Like
system
May 19, 2024, 12:55pm
8
This topic was automatically closed 7 days after the last reply. New replies are no longer allowed.