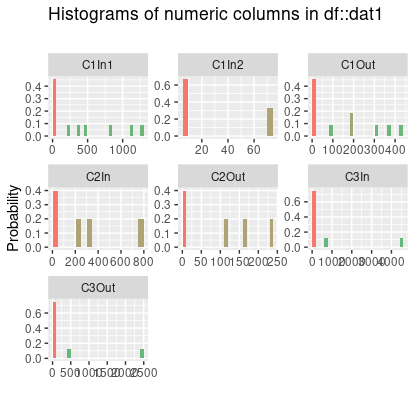
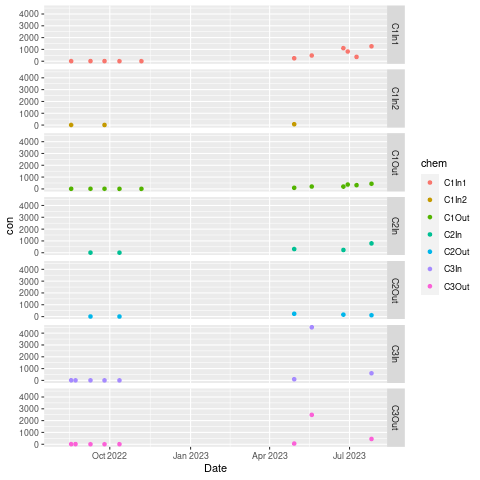
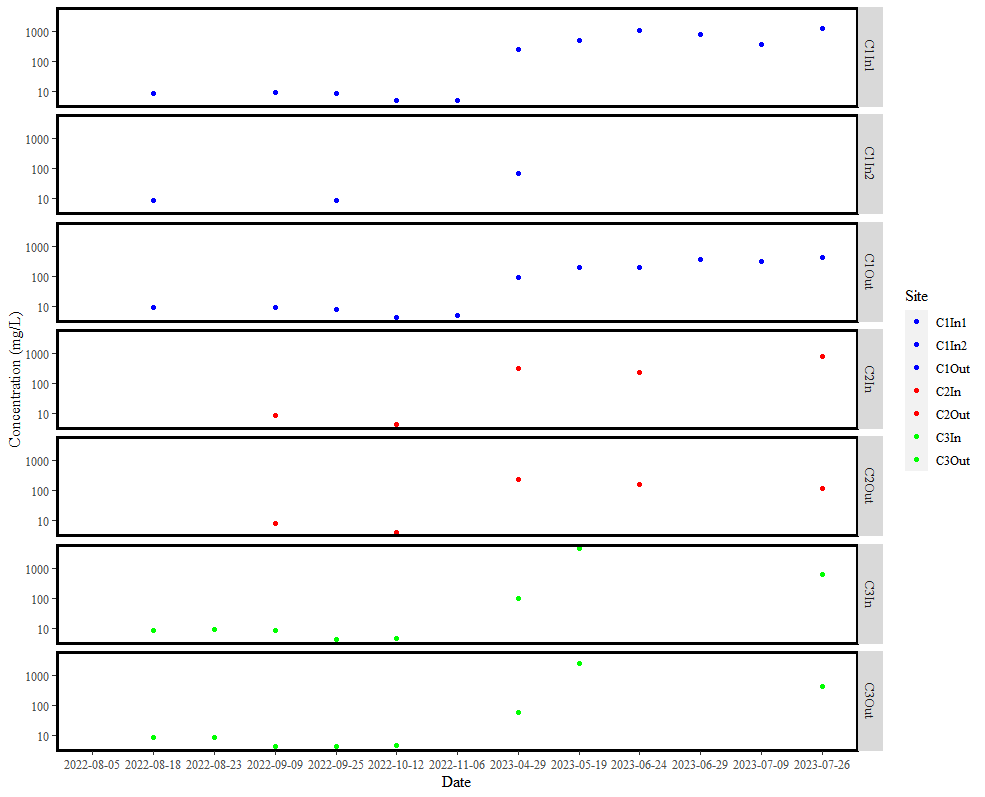
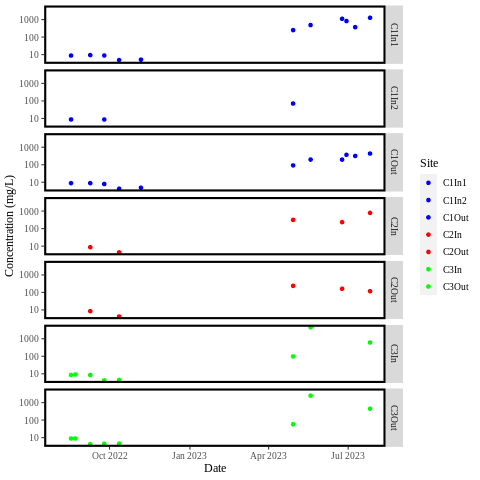
Edit:
My plot is now complete. Thanks everyone for your help. Here is my final code and plot if anyone has similar issues in the future:
library(tidyverse)
library(data.table)
library(ggplot2)
TSSdata.dat = structure(list(Date = c("2022-08-05", "2022-08-18", "2022-08-23",
"2022-09-09", "2022-09-25", "2022-10-12", "2022-11-06", "2023-04-29",
"2023-05-19", "2023-06-24", "2023-06-29", "2023-07-09", "2023-07-26"),
C1In1 = c(NA, 8.794, NA, 9.38, 8.86, 4.866, 5.124, 250, 484.63,
1107.53, 821.92, 367.5, 1265.6),
C1In2 = c(NA, 8.794, NA, NA, 8.66, NA, NA, 70.59,
NA, NA, NA, NA, NA),
C1Out = c(NA, 8.898, NA, 8.9, 7.98, 4.28, 4.88,
91.95, 197.91, 196.26, 367.92, 317.3, 433.3),
C2In = c(NA, NA, NA, 8.64, NA, 4.38, NA, 313.87, NA,
233.01, NA, NA, 788.6),
C2Out = c(NA, NA, NA, 8.5, NA, 4.21, NA, 237.7, NA,
162.16, NA, NA, 117.2),
C3In = c(NA, 8.52, 9.1, 8.5, 4.21, 4.46, NA, 98.16,
4494.04, NA, NA, NA, 606.6),
C3Out = c(NA, 8.96, 8.85, 4.23, 4.48, 4.54, NA,
57.43, 2487.91, NA, NA, NA, 447.6)),
row.names = c(NA, 13L),
class = "data.frame")
DT <-as.data.table(TSSdata.dat)
DT2 <- melt(DT, id.vars = "Date",
measure.vars = c("C1In1", "C1In2",
"C1Out", "C2In", "C2Out", "C3In", "C3Out"),
variable.name = "Site",
value.name = "con")
setDF(DT2)
p <- ggplot(DT2, aes(Date, con, colour = Site)) + geom_point() +
facet_grid(Site ~ .) + scale_y_log10() +
theme(text=element_text(family="Times New Roman", size=12)) + ylab("Concentration (mg/L)") +
scale_colour_manual(values = c("C1In1" = "blue", "C1In2" = "blue", "C1Out" = "blue",
"C2In" = "red", "C2Out" = "red", "C3In" = "green", "C3Out" = "green")) +
theme(panel.grid.minor = element_blank(),
panel.background = element_blank(),
panel.border = element_rect(colour = "black", fill = NA, size = 2))
p