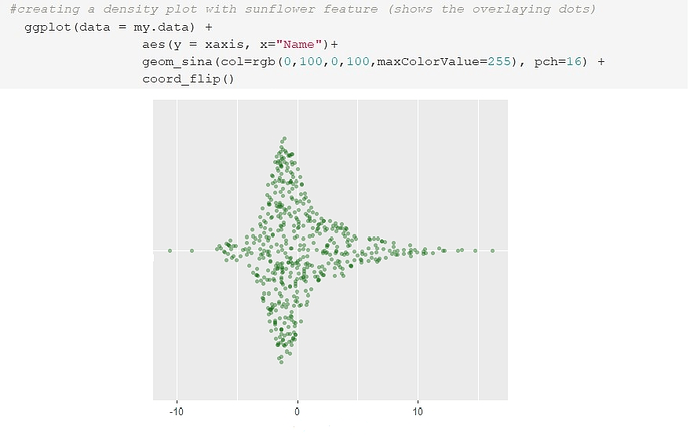
# there are several ways to approach this
# let's use the penguins data to illustrate
# install penguins data
remotes::install_github("allisonhorst/palmerpenguins")
#> Using github PAT from envvar GITHUB_PAT
#> Skipping install of 'palmerpenguins' from a github remote, the SHA1 (95e62697) has not changed since last install.
#> Use `force = TRUE` to force installation
# load packages
library(tidyverse)
library(palmerpenguins)
library(ggbeeswarm)
library(ggforce)
# peek at penguins data
glimpse(penguins)
#> Rows: 344
#> Columns: 7
#> $ species <chr> "Adelie", "Adelie", "Adelie", "…
#> $ island <chr> "Torgersen", "Torgersen", "Torg…
#> $ culmen_length_mm <dbl> 39.1, 39.5, 40.3, NA, 36.7, 39.…
#> $ culmen_depth_mm <dbl> 18.7, 17.4, 18.0, NA, 19.3, 20.…
#> $ flipper_length_mm <dbl> 181, 186, 195, NA, 193, 190, 18…
#> $ body_mass_g <dbl> 3750, 3800, 3250, NA, 3450, 365…
#> $ sex <chr> "MALE", "FEMALE", "FEMALE", NA,…
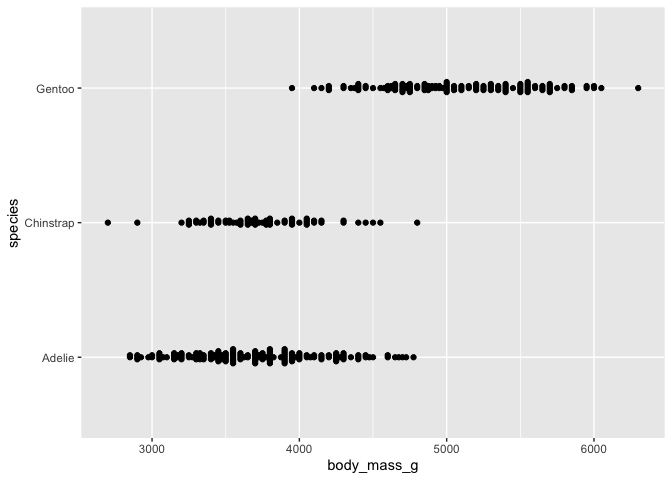
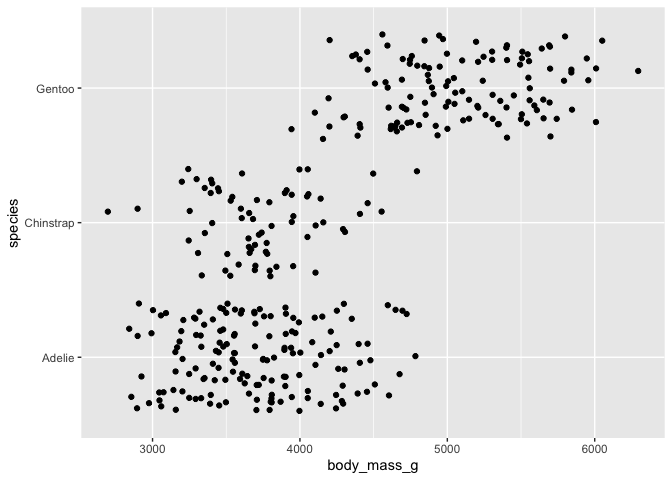
# clunky jitter version
ggplot(data = penguins) +
aes(x = body_mass_g, y = species) +
geom_jitter()
#> Warning: Removed 2 rows containing missing values
#> (geom_point).
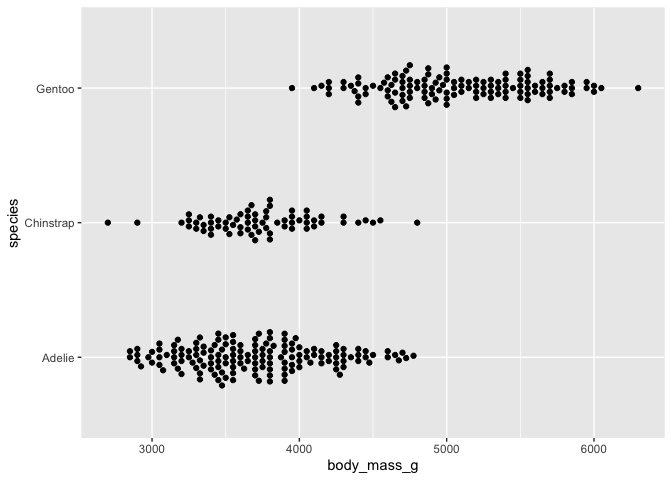
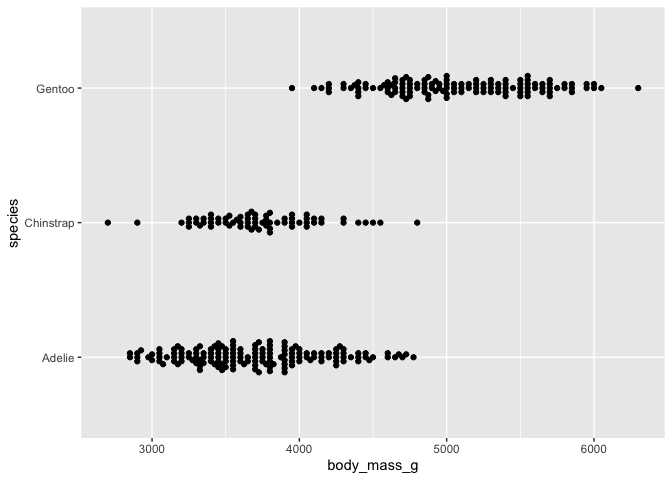
# lined up beeswarm version
ggplot(data = penguins) +
aes(y = body_mass_g, x = species) +
geom_beeswarm() +
coord_flip()
#> Warning: Removed 2 rows containing missing values
#> (position_beeswarm).
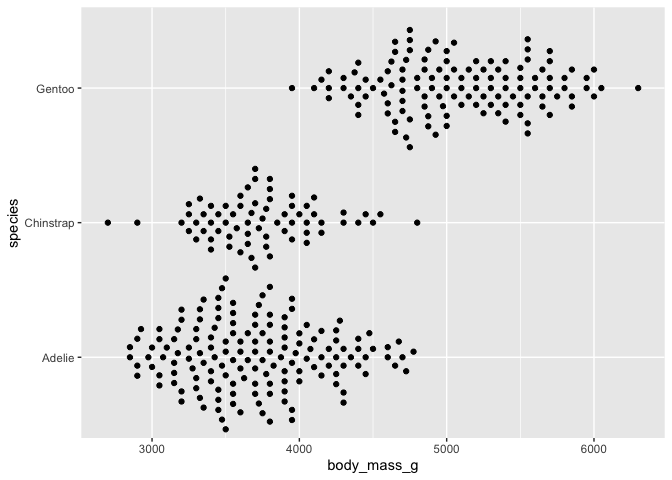
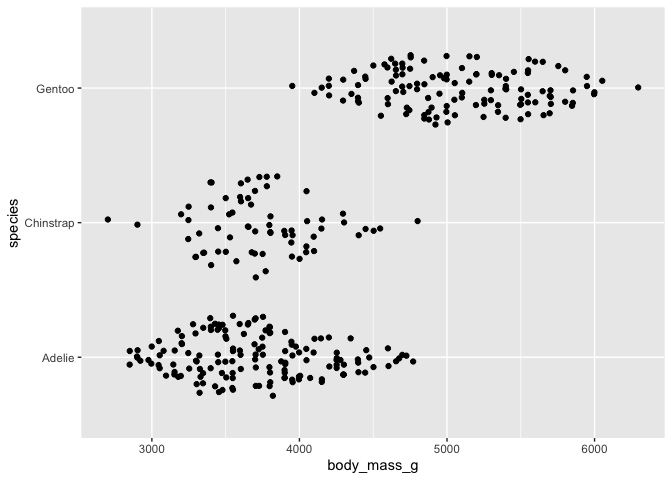
# version that corresponds to geom_violin with geom_sina
ggplot(data = penguins) +
aes(y = body_mass_g, x = species) +
geom_sina() +
coord_flip()
#> Warning: Removed 2 rows containing non-finite values
#> (stat_sina).
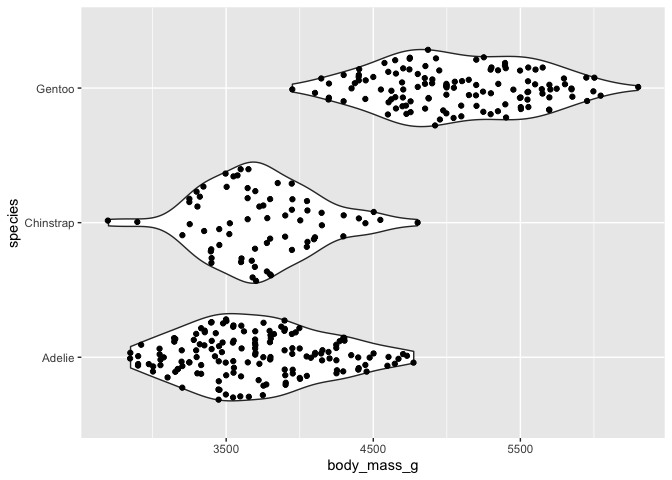
# geom_sina with geom_violin
ggplot(data = penguins) +
aes(y = body_mass_g, x = species) +
geom_violin() +
geom_sina() +
coord_flip()
#> Warning: Removed 2 rows containing non-finite values
#> (stat_ydensity).
#> Warning: Removed 2 rows containing non-finite values
#> (stat_sina).
Created on 2020-06-11 by the reprex package (v0.3.0)