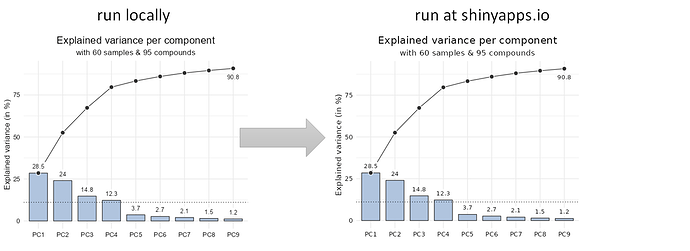
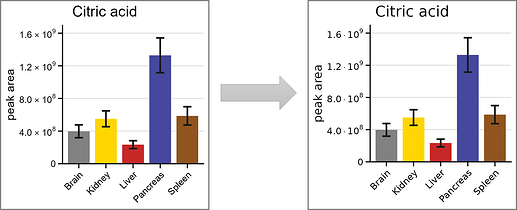
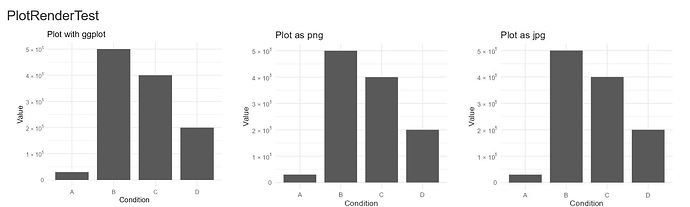
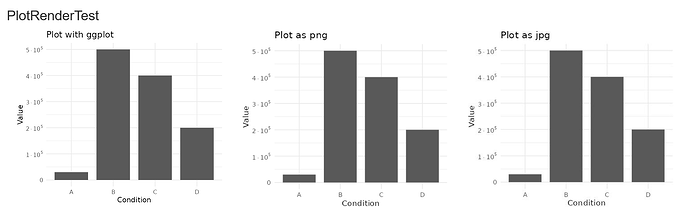
Repex:
#library(ggplot2)
library(tidyverse)
library(shiny)
# Define UI
ui <- fluidPage(
titlePanel("PlotRenderTest"),
column(width = 4, plotOutput("Plot_ggplot")),
column(width = 4, imageOutput("Plot_png") ),
column(width = 4, imageOutput("Plot_jpg") )
)
# Define server logic
server <- function(input, output) {
# Function for exp. numbers on Y-Axis
nice_scientific <- function(l) {
# turn in to character string in scientific notation
l <- format(l, scientific = TRUE)
l <- gsub("0e\\+00","0",l)
# quote the part before the exponent to keep all the digits
l <- gsub("^(.*)e", "'\\1'e", l)
# turn the 'e+' into plotmath format
l <- gsub("e\\+","e",l)
l <- gsub("e", "%*%10^", l)
# return this as an expression
parse(text=l)
}
# Data
PlotData = data.frame(Condition = c("A","B","C","D"),
Value = c(30000,500000,400000,200000))
# Base Plot
Plot = reactive({
ggplot(PlotData, aes(x = Condition, y = Value)) +
geom_col(width = 0.8) +
theme_minimal(base_size = 16) +
scale_y_continuous(labels = nice_scientific)
})
# PlotOutput
output$Plot_ggplot = renderPlot({
Plot() + labs(title = "Plot with ggplot") })
# save plot as png
output$Plot_png = renderImage({
ggsave("Plot.png",
plot = Plot() + labs(title = "Plot as png"),
bg = "white", dpi = 80, width=15, height = 13, units = "cm")
# Return the filename for the download function
list(src = "Plot.png", contentType = 'image/png')
}, deleteFile = FALSE)
# save plot as jpg
output$Plot_jpg = renderImage({
ggsave("Plot.jpg",
plot = Plot() + labs(title = "Plot as jpg"),
bg = "white", dpi = 80, width=15, height = 13, units = "cm")
# Return the filename for the download function
list(src = "Plot.jpg", contentType = 'image/jpg')
}, deleteFile = FALSE)
}
# Run the application
shinyApp(ui = ui, server = server)
sessionInfo()
R version 4.3.0 (2023-04-21 ucrt)
Platform: x86_64-w64-mingw32/x64 (64-bit)
Running under: Windows 11 x64 (build 22621)
Matrix products: default
locale:
[1] LC_COLLATE=German_Germany.utf8 LC_CTYPE=German_Germany.utf8 LC_MONETARY=German_Germany.utf8 LC_NUMERIC=C
[5] LC_TIME=German_Germany.utf8
time zone: Europe/Berlin
tzcode source: internal
attached base packages:
[1] stats graphics grDevices utils datasets methods base
other attached packages:
[1] lubridate_1.9.2 forcats_1.0.0 stringr_1.5.0 dplyr_1.1.2 purrr_1.0.1 readr_2.1.4 tidyr_1.3.0 tibble_3.2.1
[9] ggplot2_3.4.2 tidyverse_2.0.0 shiny_1.7.4
loaded via a namespace (and not attached):
[1] writexl_1.4.2 tidyselect_1.2.0 vipor_0.4.5 farver_2.1.1 fastmap_1.1.1 tweenr_2.0.2
[7] promises_1.2.0.1 shinyjs_2.1.0 digest_0.6.31 timechange_0.2.0 mime_0.12 lifecycle_1.0.3
[13] ellipsis_0.3.2 magrittr_2.0.3 compiler_4.3.0 sass_0.4.6 rlang_1.1.1 tools_4.3.0
[19] igraph_1.4.3 utf8_1.2.3 yaml_2.3.7 knitr_1.43 labeling_0.4.2 rARPACK_0.11-0
[25] plyr_1.8.8 mapproj_1.2.11 RColorBrewer_1.1-3 BiocParallel_1.34.2 withr_2.5.0 grid_4.3.0
[31] polyclip_1.10-4 fansi_1.0.4 xtable_1.8-4 colorspace_2.1-0 extrafontdb_1.0 scales_1.2.1
[37] MASS_7.3-60 pals_1.7 dichromat_2.0-0.1 cli_3.6.1 ellipse_0.4.5 rmarkdown_2.22
[43] ragg_1.2.5 generics_0.1.3 rstudioapi_0.14 RSpectra_0.16-1 tzdb_0.4.0 reshape2_1.4.4
[49] readxl_1.4.2 cachem_1.0.8 ggbeeswarm_0.7.2 ggforce_0.4.1 maps_3.4.1 parallel_4.3.0
[55] cellranger_1.1.0 matrixStats_1.0.0 vctrs_0.6.2 Matrix_1.5-4 jsonlite_1.8.5 hms_1.1.3
[61] ggrepel_0.9.3 beeswarm_0.4.0 systemfonts_1.0.4 jquerylib_0.1.4 glue_1.6.2 codetools_0.2-19
[67] mixOmics_6.24.0 cowplot_1.1.1 stringi_1.7.12 gtable_0.3.3 later_1.3.1 extrafont_0.19
[73] munsell_0.5.0 pillar_1.9.0 htmltools_0.5.5 R6_2.5.1 textshaping_0.3.6 evaluate_0.21
[79] lattice_0.21-8 memoise_2.0.1 bslib_0.5.0 corpcor_1.6.10 httpuv_1.6.9 Rcpp_1.0.10
[85] gridExtra_2.3 Rttf2pt1_1.3.12 xfun_0.39 pkgconfig_2.0.3