Hello
I wanted to install the IlluminaHumanMethylationEPICanno.ilm10b2.hg19 package(wateRmelon) of Bioconductor so i can use the readEPIC function for reading my data (.idat files). But i keep getting this error:
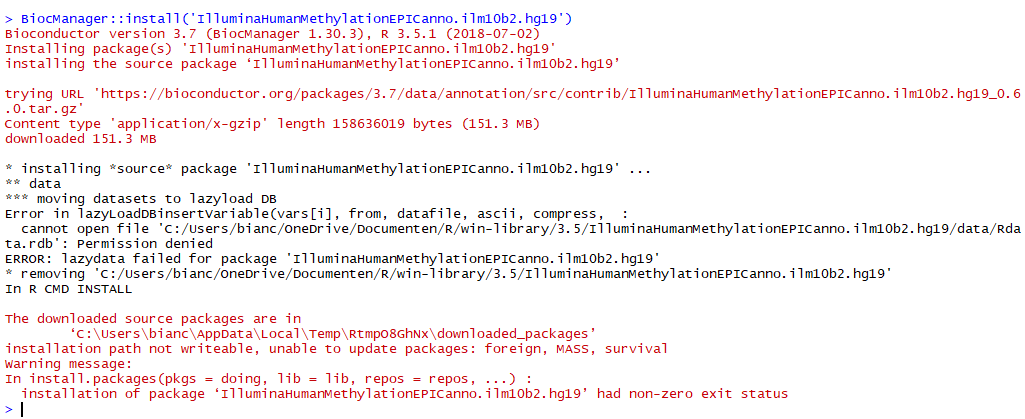
> BiocManager::install('IlluminaHumanMethylationEPICanno.ilm10b2.hg19',update=F)
Bioconductor version 3.7 (BiocManager 1.30.3), R 3.5.1 (2018-07-02)
Installing package(s) 'IlluminaHumanMethylationEPICanno.ilm10b2.hg19'
installing the source package ‘IlluminaHumanMethylationEPICanno.ilm10b2.hg19’
trying URL 'https://bioconductor.org/packages/3.7/data/annotation/src/contrib/IlluminaHumanMethylationEPICanno.ilm10b2.hg19_0.6.0.tar.gz'
Content type 'application/x-gzip' length 158636019 bytes (151.3 MB)
downloaded 151.3 MB
* installing *source* package 'IlluminaHumanMethylationEPICanno.ilm10b2.hg19' ...
** data
*** moving datasets to lazyload DB
Error in lazyLoadDBinsertVariable(vars[i], from, datafile, ascii, compress, :
cannot open file 'C:/Users/bianc/OneDrive/Documenten/R/win-library/3.5/IlluminaHumanMethylationEPICanno.ilm10b2.hg19/data/Rdata.rdb': Permission denied
ERROR: lazydata failed for package 'IlluminaHumanMethylationEPICanno.ilm10b2.hg19'
* removing 'C:/Users/bianc/OneDrive/Documenten/R/win-library/3.5/IlluminaHumanMethylationEPICanno.ilm10b2.hg19'
In R CMD INSTALL
The downloaded source packages are in
‘C:\Users\bianc\AppData\Local\Temp\RtmpioB3Cf\downloaded_packages’
Warning message:
In install.packages(pkgs = doing, lib = lib, repos = repos, ...) :
installation of package ‘IlluminaHumanMethylationEPICanno.ilm10b2.hg19’ had non-zero exit status
Does anyone know how to solve this?
Thank you!