The code you posted is indeed reproducible. But if you had pasted it within a pair of ``` (triple backticks), it would have been more readable. I did this for you now by editing your post. Hopefully, you won't mind.
Now regarding plotting mean points, note the following from the documentation of fviz_pca_ind:
Note that, fviz_pca_xxx() functions are wrapper arround the core function fviz(), whih is also a wrapper arround the function ggscatter() [in ggpubr]. Therfore, further arguments, to be passed to the function fviz() and ggscatter(), can be specified in fviz_pca_ind() and fviz_pca_var().
If you read the documentation of factoextra::fviz, you will find that there's an argument mean.point, which is TRUE by default. See below:
mean.point
logical value. If TRUE (default), group mean points are added to the plot.
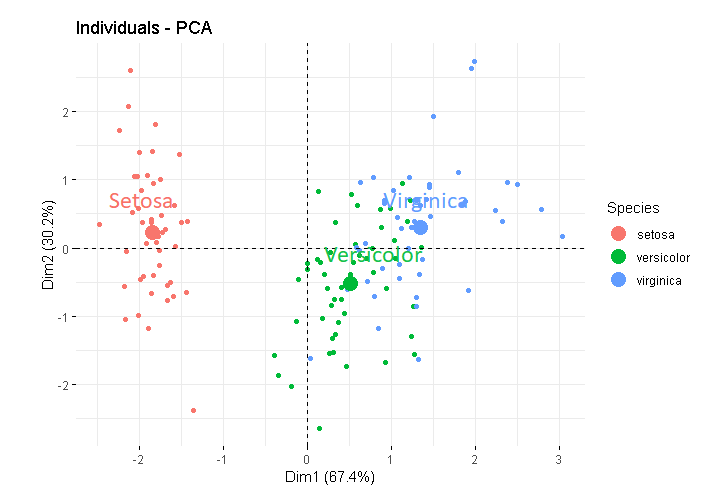
So, the plot contains the mean point by default. In the plot, you'll find that for each group, one point is a little larger than the rest and it is the mean for that group. If you explicitly set mean.point = FALSE, it'll be gone.
But it's hard (at least for me) to distinguish if there are a lot of points. In that case, you may use the mean.point.size argument of the ggpubr::ggscatter package:
mean.point.size
numeric value specifying the size of mean points.
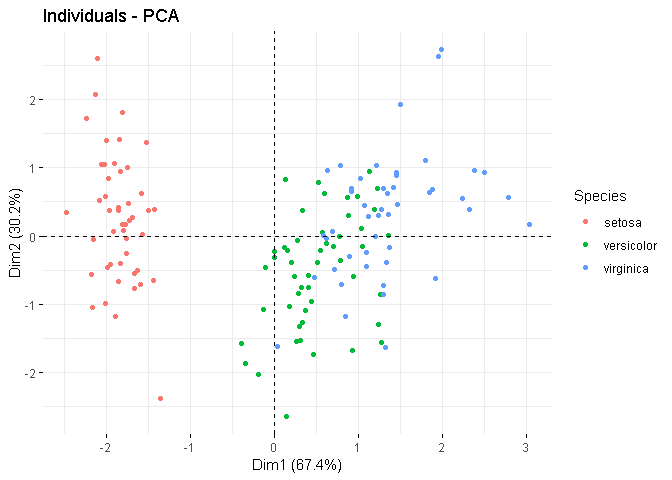
I've added an example below, where in the 1st plot, I've explicitly set the size of the mean points as 5 (so they are easy to locate), and in the 2nd plot, I skipped the mean points. Note that, the mean points, if plotted, are already coloured according to the group.
library(FactoMineR)
library(factoextra)
#> Loading required package: ggplot2
#> Welcome! Related Books: `Practical Guide To Cluster Analysis in R` at https://goo.gl/13EFCZ
res.ACP <- PCA(X = iris[, 1:3],
scale.unit = TRUE,
ncp = 5)
fviz_pca_ind(X = res.ACP,
geom.ind = c("point"),
point.size = 3,
pointshape = 16,
col.ind = iris$Species,
col = "Set1",
legend.title = "Species",
mean.point.size = 5)
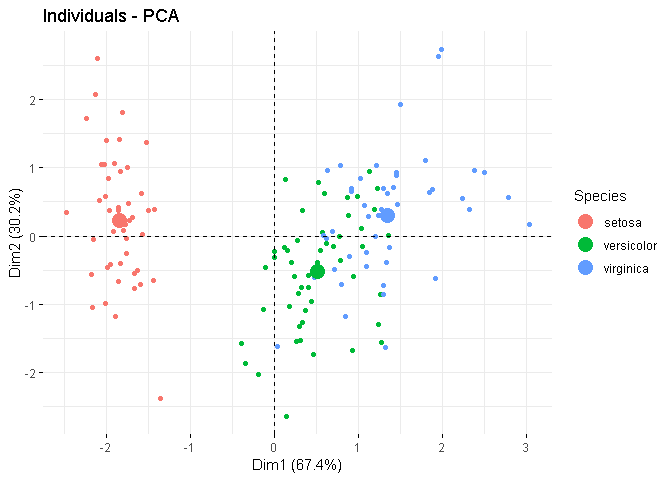
fviz_pca_ind(X = res.ACP,
geom.ind = c("point"),
point.size = 3,
pointshape = 16,
col.ind = iris$Species,
col = "Set1",
legend.title = "Species",
mean.point = FALSE)
Created on 2019-03-21 by the reprex package (v0.2.1)
Hope this helps.