I had a data set having locations as rows and species as columns.
dput(otu_table)
structure(list(Locations = c("Location 1", "Location 2", "Location 3",
"Location 4", "Location 5", "Location 6", "Location 7", "Location 8"
), Species.1 = c(1, 0, 0, 0, 0, 0, 0, 0), Species.2 = c(0, 0,
0, 0, 2, 0, 0, 0), Species.3 = c(0, 0, 0, 0, 2, 0, 0, 0), Species.4 = c(0,
0, 3, 0, 0, 1, 3, 0), Species.5 = c(0, 0, 0, 0, 0, 0, 0, 2),
Species.6 = c(0, 0, 0, 0, 0, 0, 0, 2), Species.7 = c(0, 0,
0, 0, 0, 0, 0, 2), Species.8 = c(4, 0, 0, 0, 0, 0, 0, 0),
Species.9 = c(0, 0, 0, 0, 0, 0, 0, 4), Species.10 = c(1,
0, 0, 0, 0, 0, 0, 5), Species.11 = c(0, 0, 0, 0, 3, 0, 0,
0), Species.12 = c(7, 2, 0, 0, 0, 3, 2, 0), Species.13 = c(4,
6, 0, 0, 2, 4, 1, 1), Species.14 = c(5, 0, 0, 0, 8, 0, 1,
12), Species.15 = c(0, 0, 3, 0, 0, 1, 2, 0), Species.16 = c(1,
2, 0, 0, 0, 3, 1, 2), Species.17 = c(0, 3, 5, 1, 0, 1, 1,
0), Species.18 = c(2, 0, 0, 0, 0, 0, 0, 0), Species.19 = c(2,
0, 0, 0, 0, 0, 0, 0), Species.20 = c(0, 1, 0, 1, 0, 0, 1,
1), Species.21 = c(0, 0, 2, 0, 1, 0, 2, 0), Species.22 = c(0,
3, 1, 0, 0, 3, 1, 0), Species.23 = c(0, 3, 5, 11, 0, 1, 1,
2), Species.24 = c(12L, 11L, 7L, 10L, 2L, 4L, 9L, 6L)), row.names = c(NA,
-8L), class = "data.frame")
data.pca <- prcomp(otu_table[,-1], center = TRUE, scale = TRUE)
plot_1 <- autoplot(data.pca, loadings = TRUE, loadings.label = TRUE,
data = otu_table, colour = 'Locations')
plot_1
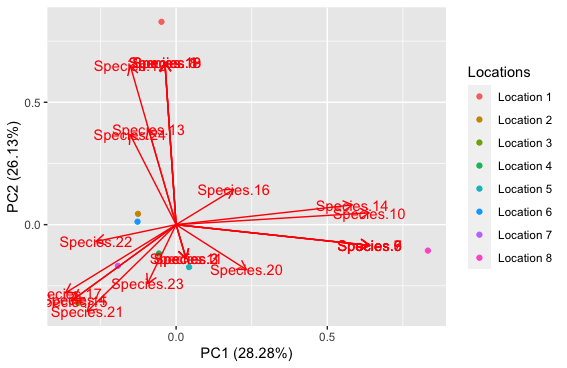
First, I want to ask that is this the correct way to read the data as I have done or does it need to be transposed?
Second, I want to see the abundance of species in all locations. So, how to add ellipses to the plot? Ellipses should be added according to species or locations? I used autoplot() function for visualisation so can i add ellipses using this autoplot() function?