Let's work with this
library(FactoMineR)
data(wine)
res <- MFA(wine, group=c(2,5,3,10,9,2), type=c("n",rep("s",5)),
ncp=5, name.group=c("orig","olf","vis","olfag","gust","ens"),
num.group.sup=c(1,6))
summary(res)
#>
#> Call:
#> MFA(base = wine, group = c(2, 5, 3, 10, 9, 2), type = c("n",
#> rep("s", 5)), ncp = 5, name.group = c("orig", "olf", "vis",
#> "olfag", "gust", "ens"), num.group.sup = c(1, 6))
#>
#>
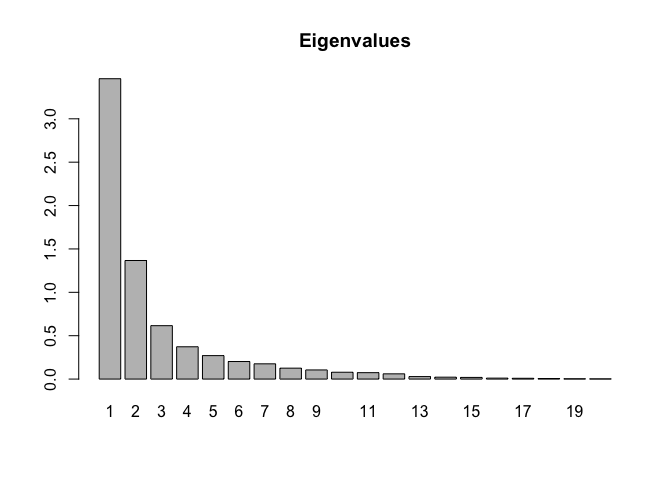
#> Eigenvalues
#> Dim.1 Dim.2 Dim.3 Dim.4 Dim.5 Dim.6 Dim.7
#> Variance 3.462 1.367 0.615 0.372 0.270 0.202 0.176
#> % of var. 49.378 19.494 8.778 5.309 3.857 2.887 2.506
#> Cumulative % of var. 49.378 68.873 77.651 82.960 86.816 89.703 92.209
#> Dim.8 Dim.9 Dim.10 Dim.11 Dim.12 Dim.13 Dim.14
#> Variance 0.126 0.105 0.079 0.074 0.060 0.029 0.022
#> % of var. 1.796 1.502 1.124 1.054 0.861 0.409 0.313
#> Cumulative % of var. 94.005 95.506 96.630 97.684 98.545 98.954 99.268
#> Dim.15 Dim.16 Dim.17 Dim.18 Dim.19 Dim.20
#> Variance 0.019 0.011 0.009 0.006 0.003 0.002
#> % of var. 0.273 0.156 0.131 0.091 0.047 0.035
#> Cumulative % of var. 99.541 99.697 99.827 99.918 99.965 100.000
#>
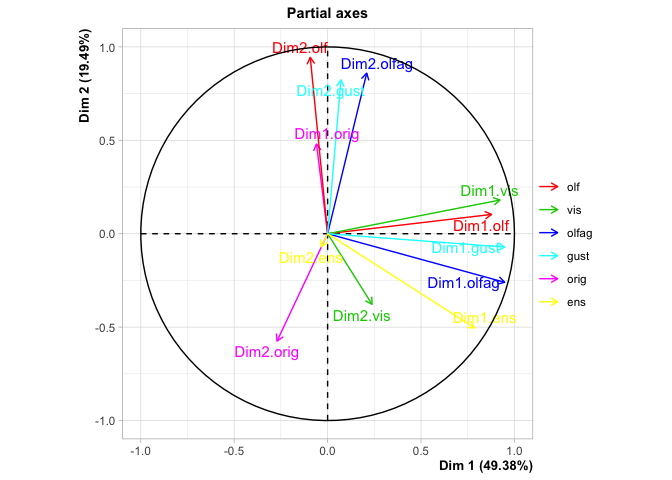
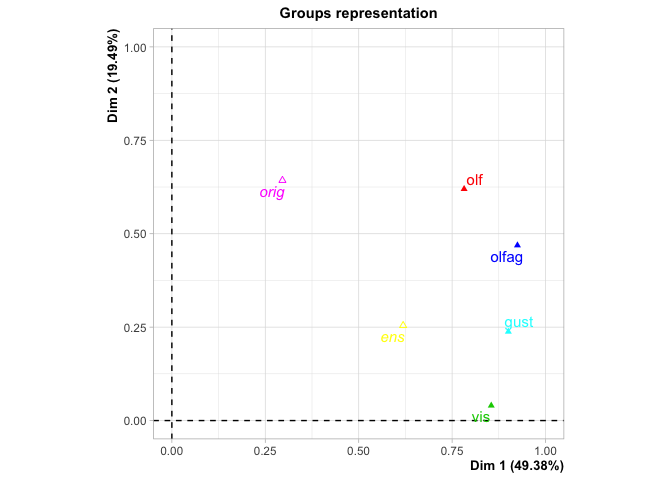
#> Groups
#> Dim.1 ctr cos2 Dim.2 ctr cos2
#> olf | 0.782 22.591 0.380 | 0.620 45.346 0.239 |
#> vis | 0.855 24.688 0.728 | 0.040 2.937 0.002 |
#> olfag | 0.925 26.712 0.625 | 0.469 34.309 0.161 |
#> gust | 0.900 26.009 0.722 | 0.238 17.408 0.050 |
#> Dim.3 ctr cos2
#> olf 0.374 60.695 0.087 |
#> vis 0.014 2.337 0.000 |
#> olfag 0.180 29.263 0.024 |
#> gust 0.047 7.705 0.002 |
#>
#> Supplementary groups
#> Dim.1 cos2 Dim.2 cos2 Dim.3 cos2
#> orig | 0.296 0.033 | 0.643 0.156 | 0.196 0.015 |
#> ens | 0.619 0.380 | 0.254 0.064 | 0.010 0.000 |
#>
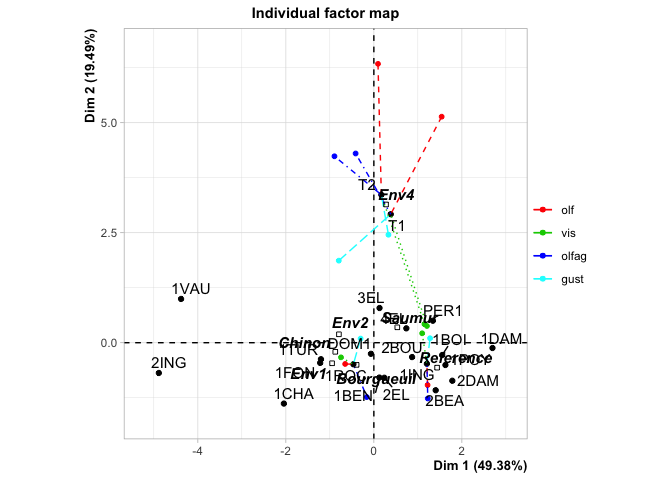
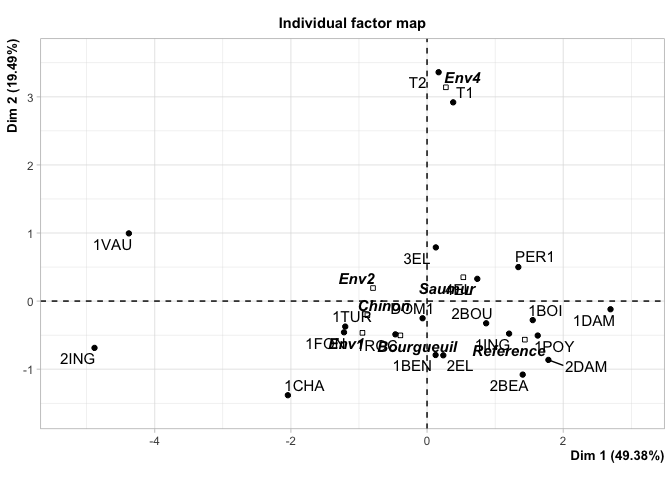
#> Individuals (the 10 first)
#> Dim.1 ctr cos2 Dim.2 ctr cos2
#> 2EL | 0.239 0.078 0.016 | -0.797 2.211 0.182 |
#> 1CHA | -2.045 5.751 0.419 | -1.383 6.667 0.192 |
#> 1FON | -1.220 2.048 0.367 | -0.459 0.734 0.052 |
#> 1VAU | -4.381 26.404 0.874 | 0.995 3.446 0.045 |
#> 1DAM | 2.696 9.996 0.754 | -0.120 0.050 0.002 |
#> 2BOU | 0.869 1.038 0.219 | -0.326 0.371 0.031 |
#> 1BOI | 1.553 3.318 0.617 | -0.280 0.272 0.020 |
#> 3EL | 0.129 0.023 0.003 | 0.789 2.167 0.115 |
#> DOM1 | -0.066 0.006 0.002 | -0.253 0.222 0.027 |
#> 1TUR | -1.202 1.987 0.310 | -0.375 0.489 0.030 |
#> Dim.3 ctr cos2
#> 2EL 0.936 6.775 0.250 |
#> 1CHA 1.514 17.725 0.229 |
#> 1FON 0.062 0.030 0.001 |
#> 1VAU -0.033 0.009 0.000 |
#> 1DAM -0.690 3.683 0.049 |
#> 2BOU 0.391 1.183 0.044 |
#> 1BOI -0.414 1.324 0.044 |
#> 3EL 1.858 26.707 0.636 |
#> DOM1 -0.459 1.629 0.090 |
#> 1TUR -0.716 3.964 0.110 |
#>
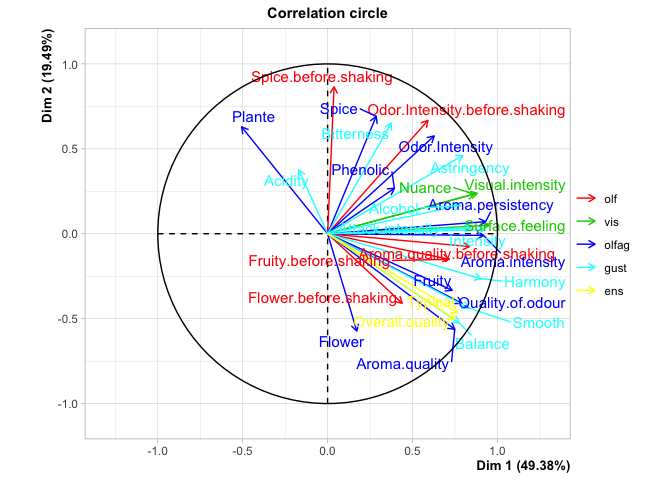
#> Continuous variables (the 10 first)
#> Dim.1 ctr cos2 Dim.2 ctr cos2
#> Odor.Intensity.before.shaking | 0.591 4.497 0.349 | 0.667 14.530 0.445 |
#> Aroma.quality.before.shaking | 0.835 8.989 0.698 | -0.075 0.186 0.006 |
#> Fruity.before.shaking | 0.716 6.606 0.513 | -0.151 0.741 0.023 |
#> Flower.before.shaking | 0.439 2.480 0.192 | -0.409 5.469 0.168 |
#> Spice.before.shaking | 0.038 0.019 0.001 | 0.865 24.420 0.748 |
#> Visual.intensity | 0.881 7.912 0.776 | 0.238 1.466 0.057 |
#> Nuance | 0.862 7.577 0.744 | 0.234 1.408 0.055 |
#> Surface.feeling | 0.950 9.198 0.903 | 0.049 0.063 0.002 |
#> Odor.Intensity | 0.627 2.416 0.393 | 0.576 5.155 0.331 |
#> Quality.of.odour | 0.791 3.844 0.626 | -0.410 2.612 0.168 |
#> Dim.3 ctr cos2
#> Odor.Intensity.before.shaking -0.023 0.039 0.001 |
#> Aroma.quality.before.shaking -0.354 9.092 0.125 |
#> Fruity.before.shaking -0.537 20.939 0.289 |
#> Flower.before.shaking 0.637 29.439 0.406 |
#> Spice.before.shaking 0.128 1.187 0.016 |
#> Visual.intensity 0.141 1.139 0.020 |
#> Nuance 0.142 1.155 0.020 |
#> Surface.feeling -0.027 0.043 0.001 |
#> Odor.Intensity 0.214 1.581 0.046 |
#> Quality.of.odour -0.221 1.684 0.049 |
#>
#> Supplementary continuous variables
#> Dim.1 cos2 Dim.2 cos2 Dim.3 cos2
#> Overall.quality | 0.747 0.558 | -0.504 0.254 | 0.130 0.017 |
#> Typical | 0.766 0.586 | -0.466 0.217 | 0.039 0.001 |
#>
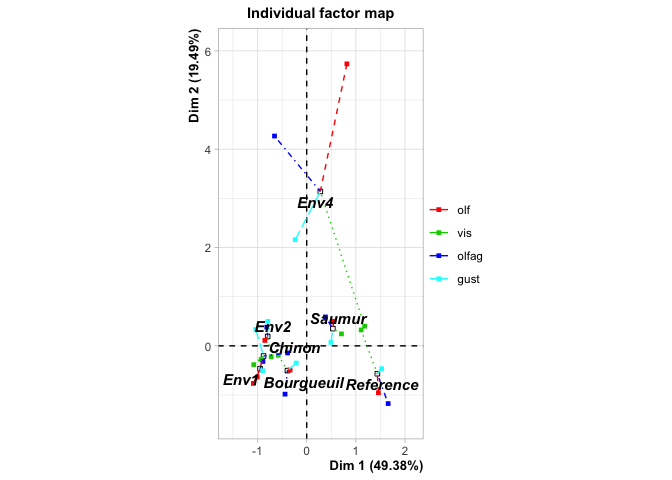
#> Supplementary categories
#> Dim.1 cos2 v.test Dim.2 cos2 v.test
#> Saumur | 0.533 0.483 1.343 | 0.350 0.209 1.405 |
#> Bourgueuil | -0.392 0.176 -0.596 | -0.504 0.291 -1.219 |
#> Chinon | -0.877 0.537 -1.022 | -0.207 0.030 -0.384 |
#> Reference | 1.437 0.823 2.442 | -0.567 0.128 -1.534 |
#> Env1 | -0.949 0.614 -1.613 | -0.467 0.149 -1.263 |
#> Env2 | -0.794 0.554 -1.067 | 0.191 0.032 0.409 |
#> Env4 | 0.277 0.008 0.216 | 3.141 0.971 3.899 |
#> Dim.3 cos2 v.test
#> Saumur 0.235 0.094 1.404 |
#> Bourgueuil -0.216 0.054 -0.780 |
#> Chinon -0.322 0.072 -0.889 |
#> Reference -0.164 0.011 -0.662 |
#> Env1 0.455 0.141 1.834 |
#> Env2 -0.382 0.129 -1.218 |
#> Env4 -0.062 0.000 -0.116 |
barplot(res$eig[,1],main="Eigenvalues",names.arg=1:nrow(res$eig))
Created on 2020-01-18 by the reprex package (v0.3.0)






