Hello everybody,
I use ggplot2 to plot 4 types of response (proboscis extension reflex test "PER") as levels A, B, C & D, as functions of 4 treatments concentrations.
My datas looks like :
'data.frame': 708 obs. of 9 variables:
Bee_Nbr : int 1204 1204 1204 1204 1206 1206 1206 1206 1102 1102 ...:
Traitement_O3: Factor w/ 4 levels "0 ppb","20 ppb",..: 1 1 1 1 1 1 1 1 1 1 ...:
Rappel : Factor w/ 4 levels "apCond","J1",..: 1 2 3 4 1 2 3 4 1 2 ...:
PER : Factor w/ 4 levels "A: CS- & NO-",..: 2 3 3 1 3 3 3 3 1 3 ...
After many hours of struggling, I managed to plot exacly the type of graph I wanted !
But, there's still a little point that miss.
I want the D response (at the botom of each sticks) to be in full black (or maybe in "grey100" if it's possible). I tried many ways to do that, but the simple "black" choice is not easy to find in ggpattern.
Can please somebody give me the solution ?
Here is my code :
PERdatas %>% filter(Rappel=="J1") %>%
ggplot(aes(Traitement_O3, fill = Traitement_O3, pattern = PER,)) +
scale_y_continuous(labels = scales::percent)+
geom_bar_pattern(position = "fill",
color = "black",
pattern_fill = "black") +
scale_fill_manual (values = c("0 ppb" = "white",
"20 ppb" ="seashell1",
"40 ppb" ="plum2",
"80 ppb" ="skyblue")) +
scale_pattern_manual(values = c("A: CS- & NO-" = "none",
"B: CS- | NO+"="stripe",
"C: CS+ & NO+"="crosshatch",
"D: CS+ | NO-"="plasma")) +
labs(y= "PER ratio (%)", x = "Concentration", title = "TEST") +
theme(plot.title = element_text(hjust = 0.5)
It works !
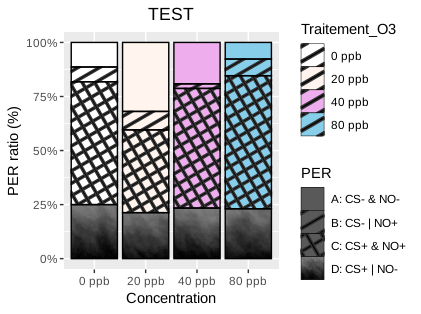
You can see I used "plasma" for the "D" pattern, because it's the given pattern that's the closest to black.
But I would really prefer the full black pattern.
Thank you for your help