Providing a reprex. See the FAQ like the following to illustrate the question will have better chances of attracting useful answers.
For example
library(vegan)
#> Loading required package: permute
#> Loading required package: lattice
#> This is vegan 2.6-4
# from help(adonis2)
data(dune)
data(dune.env)
## default test by terms
adonis2(dune ~ Management*A1, data = dune.env)
#> Permutation test for adonis under reduced model
#> Terms added sequentially (first to last)
#> Permutation: free
#> Number of permutations: 999
#>
#> adonis2(formula = dune ~ Management * A1, data = dune.env)
#> Df SumOfSqs R2 F Pr(>F)
#> Management 3 1.4686 0.34161 3.2629 0.002 **
#> A1 1 0.4409 0.10256 2.9387 0.018 *
#> Management:A1 3 0.5892 0.13705 1.3090 0.198
#> Residual 12 1.8004 0.41878
#> Total 19 4.2990 1.00000
#> ---
#> Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
## overall tests
adonis2(dune ~ Management*A1, data = dune.env, by = NULL)
#> Permutation test for adonis under reduced model
#> Permutation: free
#> Number of permutations: 999
#>
#> adonis2(formula = dune ~ Management * A1, data = dune.env, by = NULL)
#> Df SumOfSqs R2 F Pr(>F)
#> Model 7 2.4987 0.58122 2.3792 0.001 ***
#> Residual 12 1.8004 0.41878
#> Total 19 4.2990 1.00000
#> ---
#> Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
### Example of use with strata, for nested (e.g., block) designs.
dat <- expand.grid(rep=gl(2,1), NO3=factor(c(0,10)),field=gl(3,1) )
dat
#> rep NO3 field
#> 1 1 0 1
#> 2 2 0 1
#> 3 1 10 1
#> 4 2 10 1
#> 5 1 0 2
#> 6 2 0 2
#> 7 1 10 2
#> 8 2 10 2
#> 9 1 0 3
#> 10 2 0 3
#> 11 1 10 3
#> 12 2 10 3
Agropyron <- with(dat, as.numeric(field) + as.numeric(NO3)+2) +rnorm(12)/2
Schizachyrium <- with(dat, as.numeric(field) - as.numeric(NO3)+2) +rnorm(12)/2
total <- Agropyron + Schizachyrium
dotplot(total ~ NO3, dat, jitter.x=TRUE, groups=field,
type=c('p','a'), xlab="NO3", auto.key=list(columns=3, lines=TRUE) )

Y <- data.frame(Agropyron, Schizachyrium)
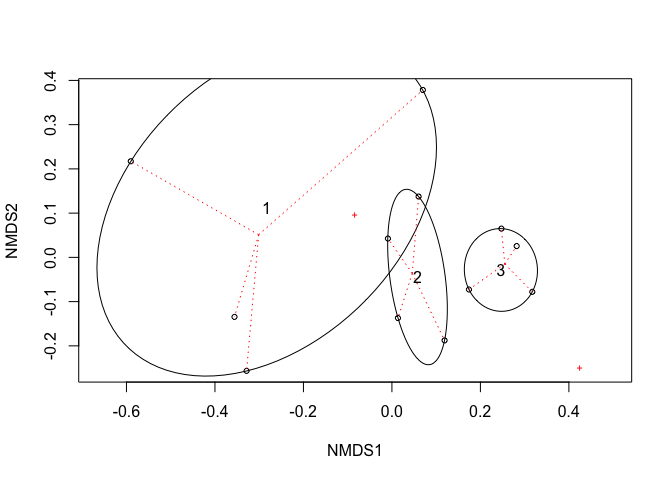
mod <- metaMDS(Y, trace = FALSE)
plot(mod)
### Ellipsoid hulls show treatment
with(dat, ordiellipse(mod, field, kind = "ehull", label = TRUE))
### Spider shows fields
with(dat, ordispider(mod, field, lty=3, col="red"))
### Incorrect (no strata)
adonis2(Y ~ NO3, data = dat, permutations = 199)
#> Permutation test for adonis under reduced model
#> Terms added sequentially (first to last)
#> Permutation: free
#> Number of permutations: 199
#>
#> adonis2(formula = Y ~ NO3, data = dat, permutations = 199)
#> Df SumOfSqs R2 F Pr(>F)
#> NO3 1 0.054321 0.17481 2.1185 0.16
#> Residual 10 0.256420 0.82519
#> Total 11 0.310741 1.00000
## Correct with strata
with(dat, adonis2(Y ~ NO3, data = dat, permutations = 199, strata = field))
#> Permutation test for adonis under reduced model
#> Terms added sequentially (first to last)
#> Blocks: strata
#> Permutation: free
#> Number of permutations: 199
#>
#> adonis2(formula = Y ~ NO3, data = dat, permutations = 199, strata = field)
#> Df SumOfSqs R2 F Pr(>F)
#> NO3 1 0.054321 0.17481 2.1185 0.01 **
#> Residual 10 0.256420 0.82519
#> Total 11 0.310741 1.00000
#> ---
#> Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Created on 2023-03-18 with reprex v2.0.2
What this illustrates that I have to guess what package is being used. I choose {vegan} because it is top hit among packages for adonis2. But the problem is that adonis2 returns a “permutational MANOVA” (formerly “nonparametric MANOVA”), not a "parametric MANOVA". Plus I am unsure how the data used compares to the data you have in mind.

