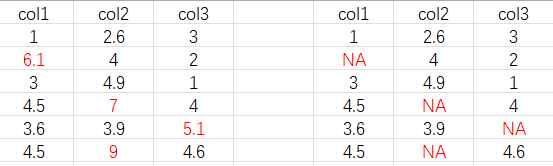
Thank you for your help.it seems very colse to what i thought.
Actually, i want to retain the data in each column that belongs to[0,5). After being selected, the data in col1 is originally is still in col1, other colums are the same. And then form a new dataframe. However, take the result "nd1"as an example. In this dataframe, there are some values that do not fall within this range [0,5). For other ranges, it seems to have the same problem.
I am very sorry that some of the words were ambiguous to you, as my English is not very good.
c5 <- data.frame(
id = c(1L,2L,3L,4L,5L,6L,7L,8L,9L,10L,
11L,12L,13L,14L,15L,16L,17L,18L,19L,20L,21L,22L,23L,
24L,25L,26L,27L,28L,29L),
col1 = c(16.6,1,10.1,8.6,8,17,2.4,7.6,5.7,
11.6,3.6,2.8,6.3,1.5,2.7,7.4,6.2,2.5,6,5.5,2,6.7,
5.6,3.5,10.4,6.8,3.5,NA,NA),
col2 = c(5.6,5.8,13.3,13.8,3,10.1,10.7,7.7,
12.3,15.8,3.5,12.7,4.2,8.1,13.6,19.8,14.3,16.8,9.9,
6.6,8.9,10.8,13.1,12.1,5.9,10.1,11.2,18.8,11),
col3 = c(9.3,10.6,1.3,2.8,5.8,NA,9.4,2.5,3.2,
7.3,12.5,18.4,15,11.8,13.7,10.2,9.5,7.6,2.5,13.6,
4.8,3.6,3.2,10.2,17.7,6.8,3.5,18.9,NA)
)
library(tidyverse)
my_rules <- list(
nd1 =function(x) x >= 0 & x<5,
nd2 =function(x) x >= 5 & x<10,
nd3 =function(x) x >= 10 & x<15,
nd4 =function(x) x >= 15 & x<20
)
(result_list <- map(my_rules,{
~mutate(c5, across(starts_with("col"),
.x,
.names = "rule_{col}"
)) |>
rowwise() |>
mutate(
rules = any(c_across(
starts_with("rule_")),
na.rm = TRUE)
) |>
filter(rules) |>
select(id,starts_with("col"))
}))
#> $nd1
#> # A tibble: 19 × 4
#> # Rowwise:
#> id col1 col2 col3
#> <int> <dbl> <dbl> <dbl>
#> 1 2 1 5.8 10.6
#> 2 3 10.1 13.3 1.3
#> 3 4 8.6 13.8 2.8
#> 4 5 8 3 5.8
#> 5 7 2.4 10.7 9.4
#> 6 8 7.6 7.7 2.5
#> 7 9 5.7 12.3 3.2
#> 8 11 3.6 3.5 12.5
#> 9 12 2.8 12.7 18.4
#> 10 13 6.3 4.2 15
#> 11 14 1.5 8.1 11.8
#> 12 15 2.7 13.6 13.7
#> 13 18 2.5 16.8 7.6
#> 14 19 6 9.9 2.5
#> 15 21 2 8.9 4.8
#> 16 22 6.7 10.8 3.6
#> 17 23 5.6 13.1 3.2
#> 18 24 3.5 12.1 10.2
#> 19 27 3.5 11.2 3.5
#>
#> $nd2
#> # A tibble: 20 × 4
#> # Rowwise:
#> id col1 col2 col3
#> <int> <dbl> <dbl> <dbl>
#> 1 1 16.6 5.6 9.3
#> 2 2 1 5.8 10.6
#> 3 4 8.6 13.8 2.8
#> 4 5 8 3 5.8
#> 5 7 2.4 10.7 9.4
#> 6 8 7.6 7.7 2.5
#> 7 9 5.7 12.3 3.2
#> 8 10 11.6 15.8 7.3
#> 9 13 6.3 4.2 15
#> 10 14 1.5 8.1 11.8
#> 11 16 7.4 19.8 10.2
#> 12 17 6.2 14.3 9.5
#> 13 18 2.5 16.8 7.6
#> 14 19 6 9.9 2.5
#> 15 20 5.5 6.6 13.6
#> 16 21 2 8.9 4.8
#> 17 22 6.7 10.8 3.6
#> 18 23 5.6 13.1 3.2
#> 19 25 10.4 5.9 17.7
#> 20 26 6.8 10.1 6.8
#>
#> $nd3
#> # A tibble: 21 × 4
#> # Rowwise:
#> id col1 col2 col3
#> <int> <dbl> <dbl> <dbl>
#> 1 2 1 5.8 10.6
#> 2 3 10.1 13.3 1.3
#> 3 4 8.6 13.8 2.8
#> 4 6 17 10.1 NA
#> 5 7 2.4 10.7 9.4
#> 6 9 5.7 12.3 3.2
#> 7 10 11.6 15.8 7.3
#> 8 11 3.6 3.5 12.5
#> 9 12 2.8 12.7 18.4
#> 10 14 1.5 8.1 11.8
#> # … with 11 more rows
#> # ℹ Use `print(n = ...)` to see more rows
#>
#> $nd4
#> # A tibble: 9 × 4
#> # Rowwise:
#> id col1 col2 col3
#> <int> <dbl> <dbl> <dbl>
#> 1 1 16.6 5.6 9.3
#> 2 6 17 10.1 NA
#> 3 10 11.6 15.8 7.3
#> 4 12 2.8 12.7 18.4
#> 5 13 6.3 4.2 15
#> 6 16 7.4 19.8 10.2
#> 7 18 2.5 16.8 7.6
#> 8 25 10.4 5.9 17.7
#> 9 28 NA 18.8 18.9
Created on 2022-10-31 with reprex v2.0.2