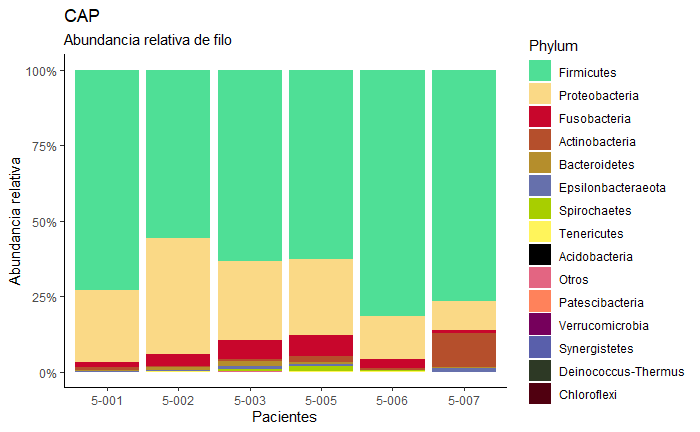
Hi, Im want make order between the most highest values, but dont find the exactly form. In this community exist other responses about this topic, but not function for my case. Im try with order() and reorder()
ggplot(d3) +
geom_bar(aes(x=Patient, y=`Relative abundance`,fill=Phylum), position="fill", stat="identity") + scale_x_discrete("Pacientes") +
scale_y_continuous("Abundancia relativa",labels=scales::percent) +
labs(title = "Grupo Neumonia",subtitle = "Abundancia relativa de filo") +
theme_classic()+ scale_fill_manual(values=c ("Grey", "Green", "Red", "Blue2", "Purple","Orange","yellow3","turquoise1", "black","green4","magenta","Purple4","Pink2","Brown","green2"))
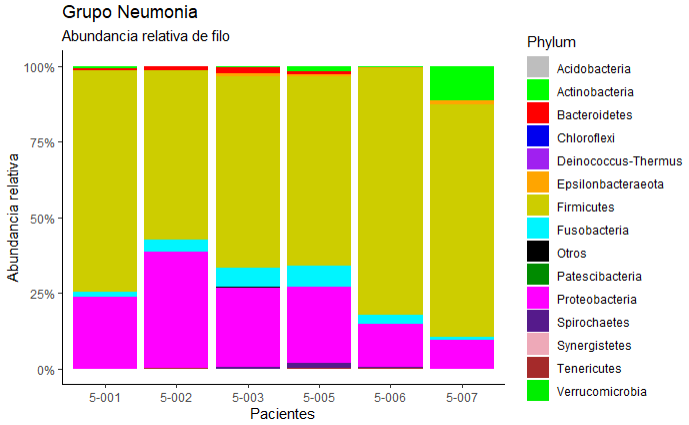
I am looking for a graph with this order, from highest to lowest in each Paciente
structure(list(Group = c("CAP", "CAP", "CAP", "CAP", "CAP", "CAP",
"CAP", "CAP", "CAP", "CAP", "CAP", "CAP", "CAP", "CAP", "CAP",
"CAP", "CAP", "CAP", "CAP", "CAP", "CAP", "CAP", "CAP", "CAP",
"CAP", "CAP", "CAP", "CAP", "CAP", "CAP", "CAP", "CAP", "CAP",
"CAP", "CAP", "CAP", "CAP", "CAP", "CAP", "CAP", "CAP", "CAP",
"CAP", "CAP", "CAP", "CAP", "CAP", "CAP", "CAP", "CAP", "CAP",
"CAP", "CAP", "CAP", "CAP", "CAP", "CAP", "CAP", "CAP", "CAP",
"CAP", "CAP", "CAP", "CAP", "CAP", "CAP", "CAP", "CAP", "CAP",
"CAP"), Phylum = structure(c(1L, 2L, 2L, 2L, 2L, 2L, 2L, 2L,
2L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 4L, 4L, 5L, 6L, 6L, 6L,
6L, 6L, 6L, 6L, 6L, 7L, 7L, 7L, 7L, 7L, 7L, 7L, 7L, 7L, 8L, 8L,
8L, 8L, 8L, 8L, 8L, 8L, 8L, 9L, 9L, 9L, 9L, 10L, 10L, 11L, 11L,
11L, 11L, 11L, 11L, 11L, 11L, 11L, 12L, 12L, 12L, 12L, 12L, 12L,
12L, 12L), .Label = c("Acidobacteria", "Actinobacteria", "Bacteroidetes",
"Chloroflexi", "Deinococcus-Thermus", "Epsilonbacteraeota", "Firmicutes",
"Fusobacteria", "Otros", "Patescibacteria", "Proteobacteria",
"Spirochaetes", "Synergistetes", "Tenericutes", "Verrucomicrobia"
), class = "factor"), `Sample type` = c(3, 3, 4, 5, 5, 5, 5,
5, 5, 3, 3, 4, 5, 5, 5, 5, 5, 5, 5, 5, 5, 3, 4, 5, 5, 5, 5, 5,
5, 3, 3, 4, 5, 5, 5, 5, 5, 5, 3, 3, 4, 5, 5, 5, 5, 5, 5, 3, 3,
4, 5, 5, 5, 3, 3, 4, 5, 5, 5, 5, 5, 5, 3, 3, 4, 5, 5, 5, 5, 5
), Patient = c("5-003", "5-002", "5-002", "5-001", "5-002", "5-003",
"5-005", "5-006", "5-007", "5-002", "5-003", "5-002", "5-001",
"5-002", "5-003", "5-005", "5-006", "5-007", "5-005", "5-006",
"5-007", "5-002", "5-002", "5-001", "5-002", "5-003", "5-005",
"5-006", "5-007", "5-002", "5-003", "5-002", "5-001", "5-002",
"5-003", "5-005", "5-006", "5-007", "5-002", "5-003", "5-002",
"5-001", "5-002", "5-003", "5-005", "5-006", "5-007", "5-002",
"5-003", "5-002", "5-001", "5-002", "5-003", "5-002", "5-003",
"5-002", "5-001", "5-002", "5-003", "5-005", "5-006", "5-007",
"5-002", "5-003", "5-002", "5-001", "5-002", "5-003", "5-005",
"5-006"), `Relative abundance` = c(0.00436022919615096, 0.00433209868520805,
0.00725767182327063, 0.034150440284692, 0.00469779532746587,
0.0144872131355983, 0.0917898572067134, 0.010858377223963, 0.322966396135543,
0.0115897705084787, 0.00185661372223202, 0.044418076778854, 0.0147403877340845,
0.00886111094701646, 0.054967018382445, 0.0321813045186884, 0.00351631386786368,
0.00601992934178261, 8.43915328287282e-05, 8.43915328287282e-05,
0.000112522043771638, 0.00261613751769058, 0.00697636671384153,
0.0065825395606408, 0.0014909170799742, 0.0318999994092593, 0.0322656960515171,
0.00241922394109021, 0.0342910928394066, 1.34899865226722, 0.332840205476504,
1.22834689083308, 2.55183116967508, 0.767653513121055, 1.70487774620597,
2.93828812900877, 1.9513291525768, 2.18799114113949, 0.0404235442249608,
0.00101269839394474, 0.187996204631464, 0.0566829795499625, 0.00998633138473284,
0.201780154993489, 0.327045320222265, 0.0708607570651888, 0.0255425039361617,
0.00210978832071821, 0.00669506160441244, 0.000393827153200732,
0.00160343912374584, 0.000225044087543275, 0.00202539678788948,
1.02566655948942, 0.140202466539461, 1.2134095895224, 0.830890901720715,
0.0496222213032922, 0.698536847734326, 1.18505403449194, 0.345245760802327,
0.273653610452623, 0.000112522043771638, 0.00123774248148801,
8.43915328287282e-05, 8.43915328287282e-05, 0.000168783065657456,
0.0162031743031158, 0.075446030348883, 0.00666693109346953)), row.names = c(NA,
-70L), groups = structure(list(Group = c("CAP", "CAP", "CAP",
"CAP", "CAP", "CAP", "CAP", "CAP", "CAP", "CAP", "CAP", "CAP",
"CAP", "CAP", "CAP", "CAP", "CAP", "CAP", "CAP", "CAP", "CAP",
"CAP", "CAP", "CAP", "CAP", "CAP", "CAP", "CAP"), Phylum = structure(c(1L,
2L, 2L, 2L, 3L, 3L, 3L, 4L, 5L, 6L, 6L, 6L, 7L, 7L, 7L, 8L, 8L,
8L, 9L, 9L, 9L, 10L, 11L, 11L, 11L, 12L, 12L, 12L), .Label = c("Acidobacteria",
"Actinobacteria", "Bacteroidetes", "Chloroflexi", "Deinococcus-Thermus",
"Epsilonbacteraeota", "Firmicutes", "Fusobacteria", "Otros",
"Patescibacteria", "Proteobacteria", "Spirochaetes", "Synergistetes",
"Tenericutes", "Verrucomicrobia"), class = "factor"), `Sample type` = c(3,
3, 4, 5, 3, 4, 5, 5, 5, 3, 4, 5, 3, 4, 5, 3, 4, 5, 3, 4, 5, 5,
3, 4, 5, 3, 4, 5), .rows = structure(list(1L, 2L, 3L, 4:9, 10:11,
12L, 13:18, 19:20, 21L, 22L, 23L, 24:29, 30:31, 32L, 33:38,
39:40, 41L, 42:47, 48:49, 50L, 51L, 52:53, 54:55, 56L, 57:62,
63:64, 65L, 66:70), ptype = integer(0), class = c("vctrs_list_of",
"vctrs_vctr", "list"))), row.names = c(NA, -28L), class = c("tbl_df",
"tbl", "data.frame"), .drop = TRUE), class = c("grouped_df",
"tbl_df", "tbl", "data.frame"))
Thanks