I'm trying to upgrade Rstudio Server from 1.2.5042 to the newest, 2021.09.1-372. But something is wrong when I try to connect with hive via odbc. Then I downgrade Rstudio Server to 1.4.1106, the error is the same.
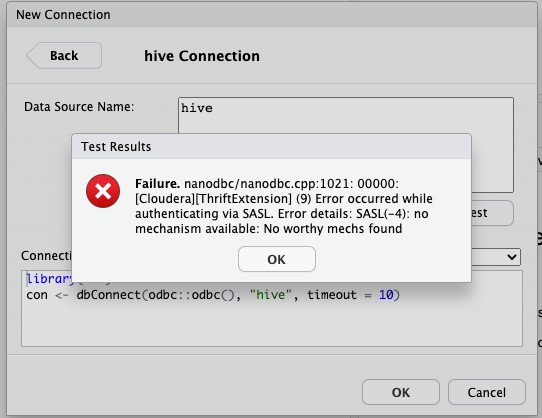
> library(DBI)
> con <- dbConnect(odbc::odbc(), "hive", timeout = 10)
Error: nanodbc/nanodbc.cpp:1021: 00000: [Cloudera][ThriftExtension] (9) Error occurred while authenticating via SASL. Error details: SASL(-4): no mechanism available: No worthy mechs found
But when I go back to 1.2.5042, odbc works fine. I compared Sys.getenv() results from Rstudio console and Rstudio terminal, there's nothing diffrent about LD_LIBRARY_PATH or PATH. I googled and found others with the same error. But they are using python or something else, and they fixed it by apt install libsasl2 libsasl2-dev or something else. But since odbc works fine in terminal both inside rstudio and outside rstudio, and in rstudio 1.2.5042, odbc env of my machine is already correct. And also, isql -v hive works fine. (DSN hive is already set in ~/.odbc.ini). I don't know how to get is work now. I use odbc to query data from hive every day. Is there any settings which should be done in new version of Rstudio Server ?
Outputs of Sys.getenv() and sessionInfo() are as followings:
> Sys.getenv()
CLICOLOR_FORCE 1
CLOUDERAHIVEINI /opt/cloudera/hiveodbc/lib/64//cloudera.hiveodbc.ini
DISPLAY :0
EDITOR vi
GIT_ASKPASS rpostback-askpass
HOME /home/rstudio
LANG en_US.UTF-8
LD_LIBRARY_PATH /usr/local/lib/R/lib::/lib:/usr/local/lib:/usr/lib/x86_64-linux-gnu:/usr/lib/jvm/java-11-openjdk-amd64/lib/server
LN_S ln -s
LOGNAME rstudio
MAKE make
MPLENGINE tkAgg
PAGER /usr/bin/pager
PATH /usr/lib/rstudio-server/bin:/usr/lib/rstudio-server/bin:/usr/local/sbin:/usr/local/bin:/usr/sbin:/usr/bin:/sbin:/bin:/usr/lib/rstudio-server/bin/postback
R_BROWSER xdg-open
R_BZIPCMD /usr/bin/bzip2
R_DOC_DIR /usr/local/lib/R/doc
R_GZIPCMD /usr/bin/gzip
R_HOME /usr/local/lib/R
R_INCLUDE_DIR /usr/local/lib/R/include
R_LIBS /usr/local/lib/R/site-library:/usr/local/lib/R/library
R_LIBS_SITE
R_LIBS_USER ~/R/x86_64-pc-linux-gnu-library/4.0
R_PAPERSIZE letter
R_PDFVIEWER /usr/bin/xdg-open
R_PLATFORM x86_64-pc-linux-gnu
R_PRINTCMD /usr/bin/lpr
R_RD4PDF times,inconsolata,hyper
R_SESSION_TMPDIR /tmp/Rtmpdxn0JW
R_SHARE_DIR /usr/local/lib/R/share
R_STRIP_SHARED_LIB strip --strip-unneeded
R_STRIP_STATIC_LIB strip --strip-debug
R_SYSTEM_ABI linux,gcc,gxx,gfortran,gfortran
R_TEXI2DVICMD /usr/bin/texi2dvi
R_UNZIPCMD /usr/bin/unzip
R_ZIPCMD /usr/bin/zip
RMARKDOWN_MATHJAX_PATH /usr/lib/rstudio-server/resources/mathjax-27
RS_RPOSTBACK_PATH /usr/lib/rstudio-server/bin/rpostback
RS_SESSION_TMP_DIR /var/run/rstudio-server/rstudio-rsession
RSTUDIO 1
RSTUDIO_CONSOLE_COLOR 256
RSTUDIO_CONSOLE_WIDTH 135
RSTUDIO_HTTP_REFERER http://10.27.5.209:8789/
RSTUDIO_PANDOC /usr/lib/rstudio-server/bin/pandoc
RSTUDIO_PROGRAM_MODE server
RSTUDIO_R_MODULE
RSTUDIO_R_PRELAUNCH_SCRIPT
RSTUDIO_R_REPO
RSTUDIO_R_VERSION_LABEL
RSTUDIO_SESSION_STREAM rstudio-d
RSTUDIO_USER_IDENTITY rstudio
RSTUDIO_USER_IDENTITY_DISPLAY rstudio
RSTUDIO_WINUTILS bin/winutils
SED /usr/bin/sed
SSH_ASKPASS rpostback-askpass
TAR /usr/bin/tar
TERM xterm-256color
USER rstudio
> sessionInfo()
R version 4.0.4 (2021-02-15)
Platform: x86_64-pc-linux-gnu (64-bit)
Running under: Ubuntu 20.04.3 LTS
Matrix products: default
BLAS/LAPACK: /usr/lib/x86_64-linux-gnu/openblas-pthread/libopenblasp-r0.3.8.so
locale:
[1] LC_CTYPE=en_US.UTF-8 LC_NUMERIC=C LC_TIME=en_US.UTF-8 LC_COLLATE=en_US.UTF-8
[5] LC_MONETARY=en_US.UTF-8 LC_MESSAGES=C LC_PAPER=en_US.UTF-8 LC_NAME=C
[9] LC_ADDRESS=C LC_TELEPHONE=C LC_MEASUREMENT=en_US.UTF-8 LC_IDENTIFICATION=C
attached base packages:
[1] stats graphics grDevices utils datasets methods base
other attached packages:
[1] DBI_1.1.1
loaded via a namespace (and not attached):
[1] bit_4.0.4 odbc_1.3.1 compiler_4.0.4 ellipsis_0.3.1 hms_1.0.0 tools_4.0.4 Rcpp_1.0.6 bit64_4.0.5
[9] vctrs_0.3.7 blob_1.2.1 lifecycle_1.0.0 pkgconfig_2.0.3 rlang_0.4.10
My odbc.ini content is as following. It works in rstudio 1.2.5042.
[hive]
Description=Cloudera ODBC Driver for Apache Hive (64-bit)
Driver=/opt/cloudera/hiveodbc/lib/64/libclouderahiveodbc64.so
Host = 192.168.90.54
Port = 10000
UID = hive_username
PWD = hive_password
AuthMech = 3
UseSASL = 1
Database = default
UseNativeQuery=1
Update, for a minimal example, we can use docker to run a hiveserver2 quickly, a docker-hive example can be found here: GitHub - big-data-europe/docker-hive. Clone it and cd to its directory and then docker-compose up -d will make a simple hive run. And then add new DSN to .odbc.ini, a new connection can be built in rstudio-server pane. Then we can make the error show again.
[hive_test]
Description=Cloudera ODBC Driver for Apache Hive (64-bit)
Driver=/opt/cloudera/hiveodbc/lib/64/libclouderahiveodbc64.so
Host = YOUR_LOCAL_IP(or localhost)
Port = 10000
Database = default
UseNativeQuery = 1
By the way, I tried Rstudio desktop on Windows and MacOS, connecting to hive both work fine. This error only occurs in Rstudio-Server. I tried to copy env variables from R in terminal to R console in Rstudio-server, nothing changes. I also tried pyodbc via Rstudio-server terminal and recticulate in Rstudio-server console, pyodbc also works fine in terminal but the same error occurs in Rstudio-server terminal.
