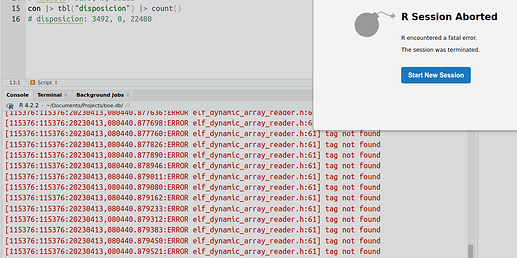
I'm using the latest RStudio desktop 2023.03 and when filling a database I'm not able to finish adding all the data.
Rstudio crashes with errors similar to this one: R Studio crashes with fatal error: elf_dynamic_array_reader.h:61] tag not found or this issue.
library("progress")
library("dplyr")
library("DBI")
library("BOE") # devtools::install_github("ropenspain/BOE")
library("duckdb")
update_sumarios <- function(con, start = "2009-01-01") {
last_date <- con |>
tbl("sumario") |>
filter(date == max(date)) |>
pull(date) |>
unique()
if (length(last_date) == 0) {
last_date <- as.Date(start)
}
date <- as.Date(last_date) + 1
days <- as.numeric(Sys.Date() - date)
pb <- progress_bar$new(total = days,
format = " downloading :what [:bar] :percent :elapsedfull :eta",
)
t.init <- Sys.time()
pb$tick(0)
while (date < Sys.Date()) {
pb$tick(tokens = list(what = date))
suma <- try(retrieve_sumario(date), silent = TRUE)
if (is(suma, "try-error")) {
date <- date + 1
next
}
suma$date <- as.character(suma$date)
dbAppendTable(con, "sumario", suma)
date <- date + 1
}
t.end <- Sys.time() - t.init
message("Updated ", days, " in ", round(t.end, 2), units(t.end))
}
create_tables <- function(con) {
types_sumario <- c(date = "DATE", sumario_nbo = "TEXT", sumario_code = "TEXT",
section = "TEXT", section_number = "TEXT", departament = "TEXT",
departament_etq = "TEXT", epigraph = "TEXT", text = "TEXT", publication = "TEXT",
pages = "DOUBLE")
types_pub <- c(identificador = "TEXT", titulo = "TEXT", diario = "TEXT",
diario_numero = "TEXT",
seccion = "TEXT", subseccion = "TEXT", departamento = "TEXT",
rango = "TEXT", numero_oficial = "TEXT", fecha_disposicion = "DATE",
fecha_publicacion = "DATE", fecha_vigencia = "DATE",
fecha_derogacion = "DATE",
letra_imagen = "TEXT", pagina_inicial = "TEXT", pagina_final = "TEXT",
suplemento_letra_imagen = "TEXT", suplemento_pagina_inicial = "TEXT",
suplemento_pagina_final = "TEXT", estatus_legislativo = "TEXT",
origen_legislativo = "TEXT", estado_consolidacion = "TEXT",
judicialmente_anulada = "TEXT",
vigencia_agotada = "TEXT", estatus_derogacion = "TEXT", url_epub = "TEXT",
url_pdf = "TEXT", url_pdf_catalan = "TEXT", url_pdf_euskera = "TEXT",
url_pdf_gallego = "TEXT", url_pdf_valenciano = "TEXT", url_eli = "TEXT",
departamento_codigo = "TEXT", fecha_actualizacion = "TIMESTAMP",
text = "TEXT")
types_an <- c(
identificador = "TEXT", titulo = "TEXT", diario = "TEXT",
diario_numero = "TEXT",
seccion = "TEXT", departamento = "TEXT", numero_anuncio = "TEXT",
fecha_publicacion = "DATE", letra_imagen = "TEXT", pagina_inicial = "TEXT",
pagina_final = "TEXT", url_pdf = "TEXT", departamento_codigo = "TEXT",
fecha_actualizacion = "TIMESTAMP", subseccion = "TEXT",
suplemento_letra_imagen = "TEXT", suplemento_pagina_inicial = "TEXT",
suplemento_pagina_final = "TEXT", text = "TEXT")
if (!dbExistsTable(con, "sumario")) {
dbCreateTable(con, name = "sumario", fields = types_sumario,
temporary = FALSE)
}
if (!dbExistsTable(con, "disposicion")) {
dbCreateTable(con, name = "disposicion", fields = types_pub,
temporary = FALSE)
}
if (!dbExistsTable(con, "anuncio")) {
dbCreateTable(con, name = "anuncio", fields = types_an, temporary = FALSE)
}
if (!dbExistsTable(con, "notas")) {
dbCreateTable(con, name = "notas",
fields = c(publication = "TEXT", codigo = "TEXT",
orden = "TEXT", text = "TEXT"), temporary = FALSE)
}
if (!dbExistsTable(con, "materias")) {
dbCreateTable(con,
name = "materias",
fields = c(publication = "TEXT", codigo = "TEXT",
orden = "TEXT", text = "TEXT"), temporary = FALSE)
}
if (!dbExistsTable(con, "alertas")) {
dbCreateTable(con,
name = "alertas",
fields = c(publication = "TEXT", codigo = "TEXT",
orden = "TEXT", text = "TEXT"), temporary = FALSE)
}
if (!dbExistsTable(con, "referencias")) {
dbCreateTable(con,
name = "referencias",
fields = c(publication = "TEXT", type = "TEXT",
referencia = "TEXT", orden = "TEXT",
codigo = "TEXT", palabra = "TEXT", texto = "TEXT"),
temporary = FALSE)
}
}
pub_collect <- function(conection) {
p <- tbl(conection, "sumario") |>
anti_join(tbl(conection, "anuncio"),
by = c(publication = "identificador")) |>
anti_join(tbl(conection, "disposicion"),
by = c(publication = "identificador")) |>
arrange(publication) |>
pull(publication)
p
}
add_anuncio <- function(con, doc) {
if (!is.list(doc$analysis[[1]])) {
return(FALSE)
}
doc <- select(doc, -analysis, -text_xml)
doc$fecha_publicacion <- as.character(doc$fecha_publicacion)
doc$fecha_actualizacion <- as.character(doc$fecha_actualizacion)
dbAppendTable(con, "anuncio", doc)
}
add_disposicion <- function(con, doc) {
redoc <- select(doc, -analysis, -text_xml)
redoc$fecha_publicacion <- as.character(redoc$fecha_publicacion)
redoc$fecha_disposicion <- as.character(redoc$fecha_disposicion)
redoc$fecha_actualizacion <- as.character(redoc$fecha_actualizacion)
redoc$fecha_derogacion <- as.character(redoc$fecha_derogacion)
redoc$fecha_vigencia <- as.character(redoc$fecha_vigencia)
dbAppendTable(con, "disposicion", redoc)
if (!is.list(doc$analysis[[1]])) {
return(FALSE)
}
if (is.list(doc$analysis[[1]]$referencias)) {
add_references(con, doc)
}
}
add_references <- function(con, doc) {
if (is.matrix(doc$analysis[[1]]$referencias$anteriores)) {
anteriores <- cbind(type = "anteriores", doc$analysis[[1]]$referencias$anteriores)
}
if (is.matrix(doc$analysis[[1]]$referencias$posteriores)) {
posteriores <- cbind(type = "posteriores", doc$analysis[[1]]$referencias$posteriores)
}
if (exists("anteriores") && exists("posteriores")) {
ref <- rbind(anteriores, posteriores)
} else if (exists("anteriores")) {
ref <- anteriores
} else if (exists("posteriores")) {
ref <- posteriores
}
if (exists("ref")) {
ref <- cbind(publication = doc$identificador, ref) %>%
as.data.frame()
dbAppendTable(con, "referencias", ref)
}
}
is.named <- function(x) {
!is.null(names(x))
}
update_publications <- function(con, publi) {
out <- anti_join(data.frame(identificador = publi),
tbl(con, "disposicion") |> select("identificador"), copy = TRUE,
by = join_by(identificador)) |>
anti_join(tbl(con, "anuncio") |> select("identificador"), copy = TRUE,
by = join_by(identificador))
publi <- out$identificador
t.init <- Sys.time()
pb <- progress_bar$new(total = length(publi),
format = " downloading :what [:bar] :percent :elapsedfull :eta",
)
pb$tick(0)
for (pub in publi) {
# Skip if already exists:
pb$tick(tokens = list(what = pub))
doc <- retrieve_document(pub)
# Empty value appearing for first time in BOE-A-2011-3164
doc$fecha_anulacion <- NULL
# message(pub)
if (strsplit(pub, "-", fixed = TRUE)[[1]][2] == "B") {
add_anuncio(con, doc)
} else {
add_disposicion(con, doc)
}
analysis <- doc$analysis[[1]]
if (!is.null(analysis) && is.named(analysis)) {
if (!is.null(analysis$notas) && is.data.frame(analysis$notas)) {
dbAppendTable(con, "notas", cbind(publication = pub, analysis$notas))
}
if (!is.null(analysis$alertas) && is.data.frame(analysis$alertas)) {
dbAppendTable(con, "alertas", cbind(publication = pub, analysis$alertas))
}
if (!is.null(analysis$materias) && is.data.frame(analysis$materias)) {
dbAppendTable(con, "materias", cbind(publication = pub, analysis$materias))
}
if (!is.null(analysis$referencias) && is.list(analysis$referencias)) {
add_references(con, doc)
}
}
}
t.end <- Sys.time() - t.init
message("Updated ", length(publi), " in ", round(t.end, 2), units(t.end))
}
clean_duplicates <- function(con, table) {
# Delete duplicated on from https://stackoverflow.com/a/8190671/2886003
str <- paste0("delete from ", table)
str2 <- paste0(" where rowid not in (select min(rowid) from ", table,
" group by ")
columns <- dbListFields(con, table)
columns <- paste0(paste0(columns, collapse = ", "), ")")
query <- paste0(str, str2, columns)
res <- dbSendStatement(con, query)
dbClearResult(res)
}
clean_con <- function(con) {
if (length(dbListTables(con)) == 0) {
create_tables(con)
res <- dbSendQuery(con, "CREATE INDEX sumario_code ON sumario (sumario_code)")
dbClearResult(res)
res <- dbSendQuery(con, "CREATE UNIQUE INDEX publication ON sumario (publication)")
dbClearResult(res)
res <- dbSendQuery(con, "CREATE UNIQUE INDEX identificador_d ON disposicion (identificador)")
dbClearResult(res)
res <- dbSendQuery(con, "CREATE UNIQUE INDEX identificador_a ON anuncio (identificador)")
dbClearResult(res)
res <- dbSendQuery(con, "CREATE INDEX publication_a ON alertas (publication)")
dbClearResult(res)
res <- dbSendQuery(con, "CREATE INDEX publication_m ON materias (publication)")
dbClearResult(res)
res <- dbSendQuery(con, "CREATE INDEX publication_n ON notas (publication)")
dbClearResult(res)
res <- dbSendQuery(con, "CREATE INDEX publication_r ON referencias (publication)")
dbClearResult(res)
} else {
clean_duplicates(con, "sumario")
clean_duplicates(con, "disposicion")
clean_duplicates(con, "anuncio")
clean_duplicates(con, "alertas")
clean_duplicates(con, "materias")
clean_duplicates(con, "notas")
clean_duplicates(con, "referencias")
}
}
And then call the functions like this:
# Update sumarios table
con <- dbConnect(duckdb::duckdb(dbdir = "boe_ddbb2.duck"))
update_sumarios(con) # Takes around 10 minutes
publications <- pub_collect(con)
# Takes around 15 minutes to hang Rstudio
update_publications(con, publications)
After 15 minutes or so adding data to the database the following message is printed in the R terminal and Rstudio crashes: ERROR elf_dynamic_array_reader.h:61] tag not found
I have tried changing the database (Originally it was MySQLite) but it didn't register entries after some time and hang up Rstudio too.
I tried to fill the database from R without RStudio but the error is similar.
Any advice on how to debug this error will be appreciated too.