Hi everyone,
I couldn't see a similar post on the forum so I haven't quoted another query/post here.
I am having some difficulty with an NMDS plot that I have created. I have six transects along with a rugosity reading and abundance data for each, as follows:
| Transect | Rugosity | Acanthastrea_brevis | Acanthastrea_ishigakiensis | Acropora_digitifera | Acropora_divaricata | Acropora_gemmifera | Acropora_latistella |
|---|---|---|---|---|---|---|---|
| ARM018 | 0.606218508 | 0 | 0 | 0 | 100 | 0 | 100 |
| ARM0115 | 0.615192661 | 25 | 100 | 50 | 0 | 0 | 0 |
| ARM068 | 0.413580544 | 0 | 0 | 0 | 0 | 0 | 0 |
| ARM0615 | 0.53388867 | 0 | 0 | 0 | 0 | 0 | 0 |
| ARM078 | 0.700824123 | 0 | 0 | 0 | 0 | 100 | 0 |
| ARM0715 | 0.768103537 | 75 | 0 | 50 | 0 | 0 | 0 |
Table 1. A sample of my data (you can see that ARM078 and ARM0715 have different data)
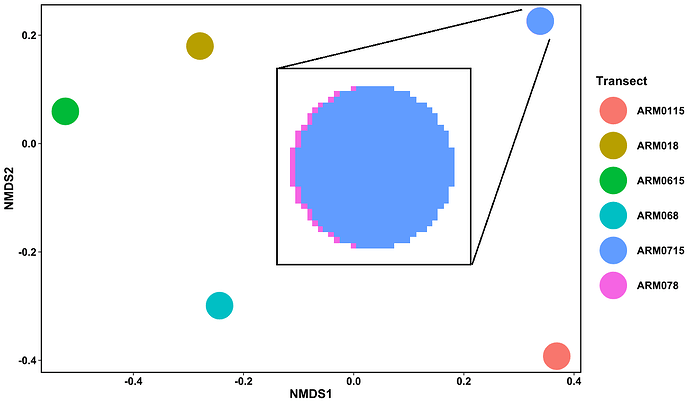
My issue is that I have 6 transects, but only 5 are showing on the NMDS plot. After some digging around to see I had generated the NMDS wrong I realised that a point could be hidden behind another so I altered the aesthetics of the plot to see if I could figure out where the missing point was. I noticed a faint pink line around the blue point:
Fig 1. My NMDS plot showing 5 points with a very zoomed in point ARM0715 (blue) showing a pink rim which could be hiding point ARM078
Non of the other points looks like this when zoomed in - they all have uniform colour. Changing the size and opacity of the points hasn't worked as the point is still hidden behind the other. Looking at Table 1 you can see that ARM078 and ARM0715 have different data so surely they shouldn't appear in exactly the same region?
My data has the shape 6 obs. of 51 variables.
Has anyone come across anything like this before? Or perhaps can think of a way for me to fix it? I though about maybe transposing the data and will try this next.
Any suggestions are very much appreciated. Thank you.
P.s. here is an outline of the nmds list object that I have generated in R:
| Name | Type | Value | ||
|---|---|---|---|---|
| nmds | "list [36] (S3: metaMDS monoMDS)" | List of length 36 | ||
| nobj | integer [1] | 6 | ||
| nfix | integer [1] | 0 | ||
| ndim | double [1] | 2 | ||
| ndis | integer [1] | 15 | ||
| ngrp | integer [1] | 1 | ||
| diss | double [15] | 0.701 0.771 0.773 0.781 0.803 0.832 ... | ||
| iidx | integer [15] | 6 4 4 3 5 5 ... | ||
| jidx | integer [15] | 5 1 3 1 2 1 ... | ||
| xinit | double [12] | -0.229 0.336 -0.205 -0.491 0.298 0.292 ... | ||
| istart | integer [1] | 1 | ||
| isform | integer [1] | 1 | ||
| ities | integer [1] | 1 | ||
| iregn | integer [1] | 1 | ||
| iscal | integer [1] | 1 | ||
| maxits | integer [1] | 200 | ||
| sratmx | double [1] | 0.999999 | ||
| strmin | double [1] | 1.00E-04 | ||
| sfgrmn | double [1] | 1.00E-07 | ||
| dist | double [15] | 0.000286 0.272546 0.454877 0.480353 0.619081 0.619261 ... | ||
| dhat | double [15] | 0.000286 0.272546 0.454877 0.480353 0.619081 0.619221 ... | ||
| points | double [6 x 2] | -0.2790 0.3687 -0.2435 -0.5235 0.3385 0.3388 0.1799 -0.3923 -0.2991 0.0594 ... | ||
| stress | double [1] | 7.60E-05 | ||
| grstress | double [1] | 7.60E-05 | ||
| iters | integer [1] | 11 | ||
| icause | integer [1] | 2 | ||
| call | language | "metaMDS(comm = m_com, distance = ""bray"")" | ||
| model | character [1] | global | ||
| distmethod | character [1] | bray | ||
| distcall | character [1] | "'vegdist(x = comm, method = distance)'" | ||
| data | character [1] | wisconsin(sqrt(m_com)) | ||
| distance | character [1] | bray | ||
| converged | double [1] | 0 | ||
| tries | double [1] | 20 | ||
| bestry | double [1] | 0 | ||
| engine | character [1] | monoMDS | ||
| species | double [49 x 2] | 0.408 0.428 0.413 -0.324 0.393 -0.324 -0.049 -0.492 -0.155 0.226 0.283 0. ... |