I downloaded my excel data sheet into R, but working with it afterwards doesnt seem to go very well. Is there someone who can help me out with showing me some basic skills? YouTube and google were not able to get me further.
Hi Daisy,
It might help if you post your code and describe what the issue is. Even post the error message. A reproducible example would be really helpful.
Hi Williaml,
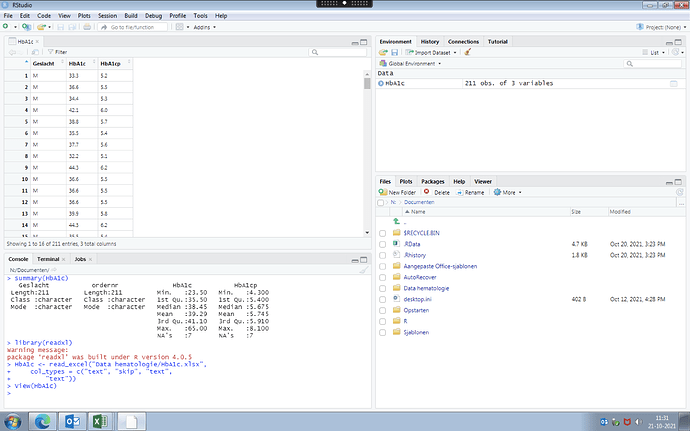
Thank you for your fast reply. I made a screenshot of what I am working on. It is a dataset where I would like to now how many values of HbA1c are >43 and how many values of HbA1cp are >6.1.
I don't know where to start and if I put in the dataset correctly (selected HbA1c and HbA1cp as numeric and Geslacht (gender) as character. Thank you very much for helping me out.
Here is an example using the iris dataset.
library(dplyr)
iris %>%
filter(Sepal.Length > 5.5) %>%
count()
You'll need to install the dplyr package if you haven't already done so. You can replace iris and Sepal.Length with the variables from your dataset.
By the way, it is best if you post the code rather than a screenshot.
Thanks again.
I downloaded the dplyr package.
HbA1c %>%
-
filter(HbA1c > 43)
Error in HbA1c %>% filter(HbA1c > 43) : could not find function "%>%"
count()
Error in count() : could not find function "count"
This is what I get when i try to add my variable.
Sorry, "%>%" is from the magrittr package.
If you want, you can just install that.
It would be better though to install the tidyverse pacakge. It contains a lot of other useful functions.
Otherwise, you can also do this:
count(filter(HbA1c, HbA1c > 43))
Thank you. I downloaded the Tidyverse package.
Now this is what I get:
HbA1c %>%
-
filter(HbA1c > 43) %>% -
count(filter(HbA1c, HbA1c > 43))
Error in HbA1c %>% filter(HbA1c > 43) %>% count(filter(HbA1c, HbA1c > :
could not find function "%>%"
count(filter(HbA1c, HbA1c > 43))
Error in count(filter(HbA1c, HbA1c > 43)) :
could not find function "count"
What happens if you run this?
library(tidyverse)
HbA1c %>%
filter(HbA1c > 43) %>%
count()
library(tidyverse)
-- Attaching packages ------------------------------------------------ tidyverse 1.3.1 --
v ggplot2 3.3.5 v purrr 0.3.4
v tibble 3.1.5 v dplyr 1.0.7
v tidyr 1.1.4 v stringr 1.4.0
v readr 2.0.2 v forcats 0.5.1
-- Conflicts --------------------------------------------------- tidyverse_conflicts() --
x dplyr::filter() masks stats::filter()
x dplyr::lag() masks stats::lag()
Warning messages:
1: package ‘tidyverse’ was built under R version 4.0.5
2: package ‘ggplot2’ was built under R version 4.0.5
3: package ‘tibble’ was built under R version 4.0.5
4: package ‘tidyr’ was built under R version 4.0.5
5: package ‘readr’ was built under R version 4.0.5
6: package ‘purrr’ was built under R version 4.0.5
7: package ‘dplyr’ was built under R version 4.0.5
8: package ‘stringr’ was built under R version 4.0.5
9: package ‘forcats’ was built under R version 4.0.5
HbA1c %>%
-
filter(HbA1c > 43) %>% -
count()
A tibble: 1 x 1
n
Thank you for your fast reply.
This happened! Does that mean the percentage is 36% of 43>?
That's the number of rows greater than 43 in your dataset (you were filtering the rows).
You could get percentage using something like this:
library(tidyverse)
filtered <- iris %>%
filter(Sepal.Length > 6) %>%
count()
filtered / count(iris) * 100
This is so exciting!!! I am so grateful for your help and finally getting somewhere.. Thank you so so much!
I got the percentages! So now a whole new question... Can I get this data visual in a graph?
library(tidyverse)
filtered <- HbA1c %>%
-
filter(HbA1c > 43) %>% -
count()
filtered / count(HbA1c) * 100
n
1 17.06161
library(tidyverse)
filtered <- HbA1c %>%
-
filter(HbA1cp > 6.1) %>% -
count()
filtered / count(HbA1c) * 100
n
1 13.27014
As a beginner to R you may benefit from studying this useful book.
https://r4ds.had.co.nz/
Particularly chapters 5 and 3
This topic was automatically closed 7 days after the last reply. New replies are no longer allowed.
If you have a query related to it or one of the replies, start a new topic and refer back with a link.