Hello everyone! New to R here ![]()
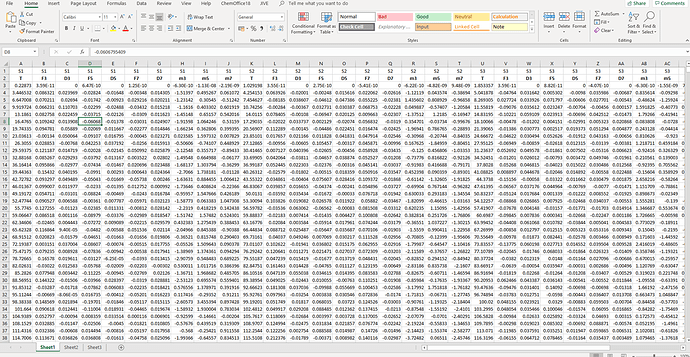
i am trying to get a tidy dataset while importing data from Excel. This is how my excel file look like:
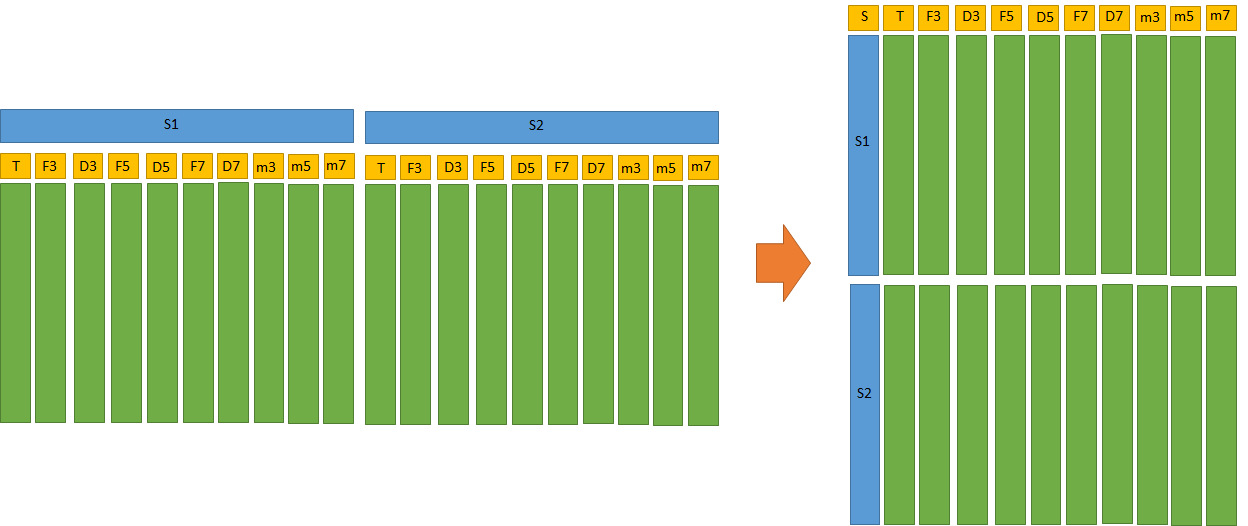
Briefly, I have 9 variables (T, F3, D3, F5, D5, F7, D7, m3, m5, m7) for 4 different sensors (S1, S2, S3, S4, first row in Excel), and I would like to have a dataset with a column named "sensor", whose observations will be S1, S1, S1.. S2 S2.. etc. and then nine more column named T, F3, D3, F5, D5, F7, D7, m3, m5, m7. I tryed to use:
dataframe_tidy <- gather(dataframe, Sensor, F3, D3, F5, D5, F7, D7, m3, m5, m7)
and I get:
Error in .f(.x[[i]], ...) : object 'F3' not found
Does someone have an idea on how I can do it?