I have made and ran the following while loop:
#this is the loop.. that you can use on the data.
replicates=dim(wideformat)[1]
toputdatain=as.data.frame(matrix(ncol=5,nrow=replicates))
names(toputdatain)=c("fly.genetic.line","pupalID","theslope","theintercept", "missing")
pupalscale=c(4,5.5,7.5,12,35,42,53,75,77,78,82,92,95,100)
p=100
while(p<(replicates+1))
{
test=wideformat[p,3:8]
datatest=as.data.frame(cbind(t(test),pupalscale))
##print(datatest)
names(datatest)[1]="timedays"
datatest$timedays= datatest$timedays - min(datatest$timedays, na.rm = T)
#HERE
#datatest$timedays <- datatest$timedays - 34 #(was 40 initially not 34)
#so maybe we need to correct he intercept here (make this equal, will it bias the slope?)
theslope=coef(lm(datatest$pupalscale~datatest$timedays))[[2]]
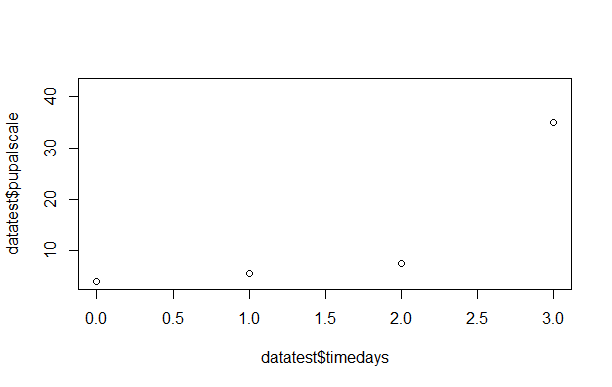
print(plot(datatest$pupalscale~datatest$timedays))
missing = length(which(is.na(datatest$timedays)== T))
theintercept=coef(lm(datatest$pupalscale~datatest$timedays))[[1]] - min(toputdatain_pupal_case_min[,3], na.rm = T)
toputdatain[p,]=cbind(wideformat[p,c(1,2)],theslope,theintercept, missing)
p=p+1
}
however when the plots appear the y axis only seems to go up to 40 rather than 100 seen on the pupal scale. any suggestions on how to fix this would be appreciated.
picture example: