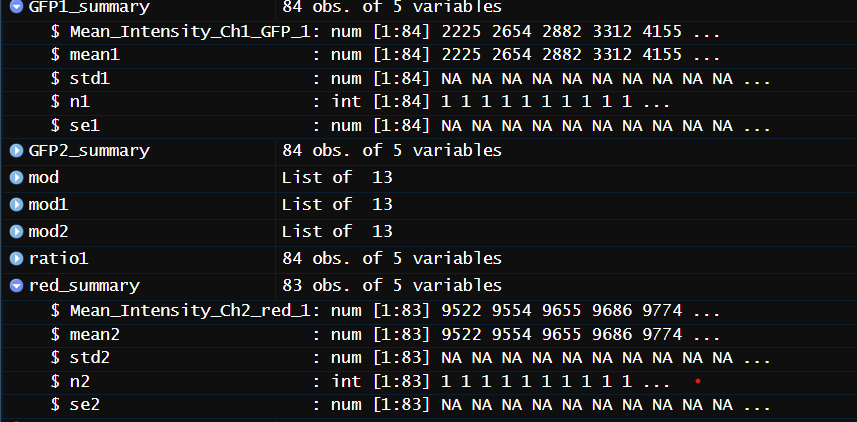
im using summary() to find the mean, se, std and length of the column, im sure there is a more efficient way to do this but thats just how i was taught
the output of dput(GFP1_summary) is
structure(list(Mean_Intensity_Ch1_GFP_1 = c(2225.2, 2654.3, 2882.3,
3312.1, 4154.9, 4596.2, 4611.3, 4734.3, 4949.5, 4954.4, 5114.7,
5125.2, 5140.7, 5275.5, 5316.2, 5655.1, 5703.6, 5841.5, 5926.7,
5936.5, 6042.6, 6306.2, 6539.8, 6565, 6599.1, 6649, 6751.3, 6784.5,
6856.8, 6859.2, 7073.3, 7101, 7153.8, 7169.1, 7242.4, 7248.4,
7253.2, 7293, 7345.7, 7378.4, 7391, 7441.6, 7462, 7517.7, 7519.7,
7521.4, 7572.4, 7600.8, 7606.7, 7628.4, 7628.6, 7637.1, 7647.8,
7680.4, 7735.5, 7749.2, 7750.2, 7762.1, 7764.8, 7773.2, 7775.2,
7784.5, 7829.6, 7936.5, 8047.3, 8056.5, 8095, 8130.3, 8140.3,
8149.5, 8158.8, 8175.2, 8183.7, 8266.1, 8268.8, 8353, 8374.6,
8388.6, 8395.5, 8422.6, 8598.5, 8676.4, 13497.8, 16393.9), mean1 = c(2225.2,
2654.3, 2882.3, 3312.1, 4154.9, 4596.2, 4611.3, 4734.3, 4949.5,
4954.4, 5114.7, 5125.2, 5140.7, 5275.5, 5316.2, 5655.1, 5703.6,
5841.5, 5926.7, 5936.5, 6042.6, 6306.2, 6539.8, 6565, 6599.1,
6649, 6751.3, 6784.5, 6856.8, 6859.2, 7073.3, 7101, 7153.8, 7169.1,
7242.4, 7248.4, 7253.2, 7293, 7345.7, 7378.4, 7391, 7441.6, 7462,
7517.7, 7519.7, 7521.4, 7572.4, 7600.8, 7606.7, 7628.4, 7628.6,
7637.1, 7647.8, 7680.4, 7735.5, 7749.2, 7750.2, 7762.1, 7764.8,
7773.2, 7775.2, 7784.5, 7829.6, 7936.5, 8047.3, 8056.5, 8095,
8130.3, 8140.3, 8149.5, 8158.8, 8175.2, 8183.7, 8266.1, 8268.8,
8353, 8374.6, 8388.6, 8395.5, 8422.6, 8598.5, 8676.4, 13497.8,
16393.9), std1 = c(NA_real_, NA_real_, NA_real_, NA_real_, NA_real_,
NA_real_, NA_real_, NA_real_, NA_real_, NA_real_, NA_real_, NA_real_,
NA_real_, NA_real_, NA_real_, NA_real_, NA_real_, NA_real_, NA_real_,
NA_real_, NA_real_, NA_real_, NA_real_, NA_real_, NA_real_, NA_real_,
NA_real_, NA_real_, NA_real_, NA_real_, NA_real_, NA_real_, NA_real_,
NA_real_, NA_real_, NA_real_, NA_real_, NA_real_, NA_real_, NA_real_,
NA_real_, NA_real_, NA_real_, NA_real_, NA_real_, NA_real_, NA_real_,
NA_real_, NA_real_, NA_real_, NA_real_, NA_real_, NA_real_, NA_real_,
NA_real_, NA_real_, NA_real_, NA_real_, NA_real_, NA_real_, NA_real_,
NA_real_, NA_real_, NA_real_, NA_real_, NA_real_, NA_real_, NA_real_,
NA_real_, NA_real_, NA_real_, NA_real_, NA_real_, NA_real_, NA_real_,
NA_real_, NA_real_, NA_real_, NA_real_, NA_real_, NA_real_, NA_real_,
NA_real_, NA_real_), n1 = c(1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L,
1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L,
1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L,
1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L,
1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L,
1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L), se1 = c(NA_real_,
NA_real_, NA_real_, NA_real_, NA_real_, NA_real_, NA_real_, NA_real_,
NA_real_, NA_real_, NA_real_, NA_real_, NA_real_, NA_real_, NA_real_,
NA_real_, NA_real_, NA_real_, NA_real_, NA_real_, NA_real_, NA_real_,
NA_real_, NA_real_, NA_real_, NA_real_, NA_real_, NA_real_, NA_real_,
NA_real_, NA_real_, NA_real_, NA_real_, NA_real_, NA_real_, NA_real_,
NA_real_, NA_real_, NA_real_, NA_real_, NA_real_, NA_real_, NA_real_,
NA_real_, NA_real_, NA_real_, NA_real_, NA_real_, NA_real_, NA_real_,
NA_real_, NA_real_, NA_real_, NA_real_, NA_real_, NA_real_, NA_real_,
NA_real_, NA_real_, NA_real_, NA_real_, NA_real_, NA_real_, NA_real_,
NA_real_, NA_real_, NA_real_, NA_real_, NA_real_, NA_real_, NA_real_,
NA_real_, NA_real_, NA_real_, NA_real_, NA_real_, NA_real_, NA_real_,
NA_real_, NA_real_, NA_real_, NA_real_, NA_real_, NA_real_)), class = c("tbl_df",
"tbl", "data.frame"), row.names = c(NA, -84L))
and the output of dput(red_summary) is
structure(list(Mean_Intensity_Ch2_red_1 = c(9521.5, 9553.9, 9654.8,
9686.5, 9774.3, 9778.7, 9804.8, 9848.2, 9858.2, 9859.9, 9865.9,
9883.6, 9909.1, 9988.3, 9990.1, 9994.5, 9997.7, 9998, 10005.3,
10017.3, 10019.8, 10045.6, 10052.7, 10102, 10110.2, 10114, 10136.4,
10170, 10179.3, 10203.7, 10218.3, 10218.6, 10239.9, 10297.2,
10348.4, 10358.9, 10385.4, 10391, 10420.5, 10446.3, 10552.8,
10570.7, 10575.7, 10598.4, 10600.5, 10740.4, 10766.8, 10789.3,
10814.9, 10902.3, 11030.7, 11059.4, 11068.8, 11091.5, 11106.1,
11130.6, 11184.6, 11195.4, 11288.1, 11299.9, 11331.6, 11380.8,
11403.6, 11601.1, 11619.9, 11774.3, 11981.9, 12043.2, 12631,
13012.7, 13571.3, 13873.9, 13903.7, 15110.4, 15758.3, 16500,
16742, 17518, 18719.2, 24637.3, 25117.6, 26339, 55895.5), mean2 = c(9521.5,
9553.9, 9654.8, 9686.5, 9774.3, 9778.7, 9804.8, 9848.2, 9858.2,
9859.9, 9865.9, 9883.6, 9909.1, 9988.3, 9990.1, 9994.5, 9997.7,
9998, 10005.3, 10017.3, 10019.8, 10045.6, 10052.7, 10102, 10110.2,
10114, 10136.4, 10170, 10179.3, 10203.7, 10218.3, 10218.6, 10239.9,
10297.2, 10348.4, 10358.9, 10385.4, 10391, 10420.5, 10446.3,
10552.8, 10570.7, 10575.7, 10598.4, 10600.5, 10740.4, 10766.8,
10789.3, 10814.9, 10902.3, 11030.7, 11059.4, 11068.8, 11091.5,
11106.1, 11130.6, 11184.6, 11195.4, 11288.1, 11299.9, 11331.6,
11380.8, 11403.6, 11601.1, 11619.9, 11774.3, 11981.9, 12043.2,
12631, 13012.7, 13571.3, 13873.9, 13903.7, 15110.4, 15758.3,
16500, 16742, 17518, 18719.2, 24637.3, 25117.6, 26339, 55895.5
), std2 = c(NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA,
NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA,
NA, NA, 0, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA,
NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA,
NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA,
NA, NA, NA, NA, NA, NA), n2 = c(1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L,
1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L,
1L, 1L, 1L, 1L, 1L, 1L, 1L, 2L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L,
1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L,
1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L,
1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L), se2 = c(NA, NA,
NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA,
NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, 0, NA, NA,
NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA,
NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA,
NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA,
NA)), class = c("tbl_df", "tbl", "data.frame"), row.names = c(NA,
-83L))