I am using mlr to create ML models for research. I would use mlr3, but the paper that I am building on uses mlr, hence why I am sticking with the older library.
I am having an issue with the performance metrics of the hyperparameter tuning on two of the models I'm using - neuralnet and randomforest.
Here is my code for a function that I use to return a predictor of a given type (NB: TUNEITERS = 100L and RESAMPLING = cv5:
getPredictor <- function(ml_alg_id,
data,
data_id,
target,
target_values) {
# Task for classification.
data.task = makeClassifTask(id = data_id,
data = data,
target = target,
positive = target_values[POS_CLV_INDEX])
# Initialise parallelisation.
parallelMap::parallelStartSocket(parallel::detectCores(), level = "mlr.tuneParams")
# Choose & train the model and set the predictor.
pred = NULL
if (ml_alg_id == NN_ALG_ID) {
# Learner: Neural network.
lrn = makeLearner("classif.nnet",
predict.type = "prob",
fix.factors.prediction = TRUE)
# Normalisation/dummy encode.
data.lrn = cpoScale() %>>% cpoDummyEncode() %>>% lrn
# Parameters for tuning.
param_grid = makeParamSet(
makeNumericParam("size", lower = 1, upper = 20),
makeNumericParam("decay", lower = 0.1, upper = 0.9)
)
# Random search for tuning method.
tune_control = makeTuneControlRandom(maxit = TUNEITERS)
# Tune.
data.lrn.tuned = tuneParams(data.lrn,
task = data.task,
resampling = RESAMPLING,
par.set = param_grid,
control = tune_control)
# Train the model.
data.model = mlr::train(data.lrn.tuned$learner, data.task)
# Set as predictor.
pred = Predictor$new(model = data.model,
data = data,
class = target_values[POS_CLV_INDEX])
}
else if (ml_alg_id == RF_ALG_ID) {
# Learner: Random Forest.
lrn = makeLearner("classif.randomForest",
predict.type = "prob",
fix.factors.prediction = TRUE)
# Parameters for tuning.
param_grid = makeParamSet(
makeIntegerParam("ntree", lower = 50, upper = 500),
makeIntegerParam("mtry", lower = 1, upper = ncol(data) - 1)
)
# Random search for tuning method.
tune_control = makeTuneControlRandom(maxit = TUNEITERS)
# Tune.
lrn.tuned = tuneParams(lrn,
task = data.task,
resampling = RESAMPLING,
par.set = param_grid,
control = tune_control)
# Train the model.
data.model = mlr::train(lrn.tuned$learner, data.task)
# Set as predictor.
pred = Predictor$new(model = data.model,
data = data,
class = target_values[POS_CLV_INDEX])
}
else if (ml_alg_id == SVM_ALG_ID) {
# Learner: Support Vector Machine.
lrn = makeLearner("classif.svm", predict.type = "prob")
# Normalisation/dummy encode.
data.lrn = cpoScale() %>>% cpoDummyEncode() %>>% lrn
# Parameters for tuning.
param.set = pSS(
cost: numeric[0.01, 1]
)
# Tune.
ctrl = makeTuneControlRandom(maxit = TUNEITERS * length(param.set$pars))
lrn.tuning = makeTuneWrapper(lrn, RESAMPLING, list(mlr::acc), param.set, ctrl, show.info = FALSE)
res = tuneParams(lrn, data.task, RESAMPLING, par.set = param.set, control = ctrl,
show.info = FALSE)
performance = resample(lrn.tuning, data.task, RESAMPLING, list(mlr::acc))$aggr
data.lrn = setHyperPars2(data.lrn, res$x)
# Train the model.
data.model = mlr::train(data.lrn, data.task)
# Set as predictor.
pred = Predictor$new(model = data.model,
data = data,
class = target_values[POS_CLV_INDEX],
conditional = FALSE)
# Fit conditional inference trees.
ctr = partykit::ctree_control(maxdepth = 5L)
set.seed(1234)
pred$conditionals = fit_conditionals(pred$data$get.x(), ctrl = ctr)
}
else {
stop("Error: Invalid ML algorithm ID passed to getPredictor()")
}
# Stop parallelisation.
parallelMap::parallelStop()
return(pred)
}
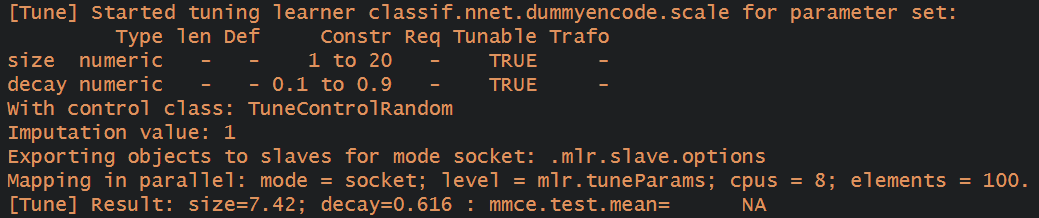
Here is the message I receive for the result of a neuralnet hyperparameter tuning:
I also get mmce.test.mean=NA for random forests too.
As shown, I am using dummy encoding with mlrCPO for the neuralnet model. I am using no such encoding for random forest as I believe it can handle heterogeneous datasets.
What am I doing wrong here to cause mmce.test.mean=NA?