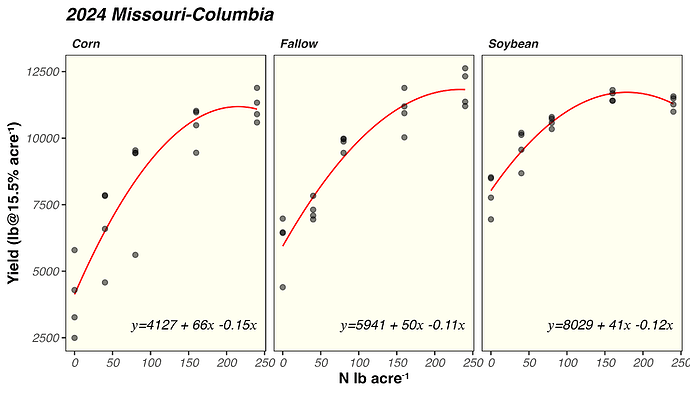
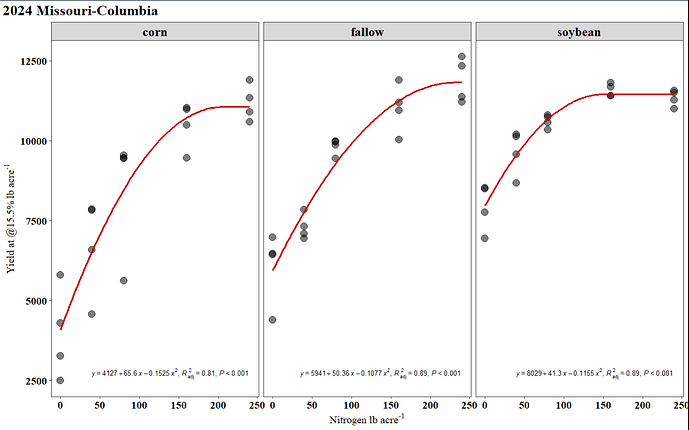
Hi all, I am currently working on a report and I have been struggling on how to extract estimate from a quadratic model and plot them on ggplot2 just as the plot in which I attached below. I will really appreciate it if someone can help me out. I have attached my sample data and code in the reprex script, along with a screenshot of expected output. Also, if I have new data, how do I get the predicted yield using the predict function? I keep running into issues here when I try. Thank you very much!
Data
data <- structure(
list(
Rep = structure(
c(
1L,
1L,
1L,
1L,
1L,
1L,
1L,
1L,
1L,
1L,
1L,
1L,
1L,
1L,
1L,
2L,
2L,
2L,
2L,
2L,
2L,
2L,
2L,
2L,
2L,
2L,
2L,
2L,
2L,
2L,
3L,
3L,
3L,
3L,
3L,
3L,
3L,
3L,
3L,
3L,
3L,
3L,
3L,
3L,
3L,
4L,
4L,
4L,
4L,
4L,
4L,
4L,
4L,
4L,
4L,
4L,
4L,
4L,
4L,
4L
),
levels = c("1", "2", "3", "4"),
class = "factor"
),
History = structure(
c(
1L,
1L,
1L,
1L,
1L,
3L,
3L,
3L,
3L,
3L,
2L,
2L,
2L,
2L,
2L,
2L,
2L,
2L,
2L,
2L,
1L,
1L,
1L,
1L,
1L,
3L,
3L,
3L,
3L,
3L,
3L,
3L,
3L,
3L,
3L,
2L,
2L,
2L,
2L,
2L,
1L,
1L,
1L,
1L,
1L,
1L,
1L,
1L,
1L,
1L,
3L,
3L,
3L,
3L,
3L,
2L,
2L,
2L,
2L,
2L
),
levels = c("corn", "fallow", "soybean"),
class = "factor"
),
N rate = c(
160,
240,
80,
0,
40,
240,
80,
160,
0,
40,
0,
160,
240,
40,
80,
0,
40,
160,
80,
240,
0,
80,
40,
160,
240,
80,
240,
160,
40,
0,
240,
0,
40,
160,
80,
160,
80,
0,
40,
240,
40,
240,
80,
0,
160,
80,
240,
0,
40,
160,
80,
240,
0,
160,
40,
40,
240,
0,
160,
80
),
Yield (lbs/ac) = c(
10985.3700627973,
10626.7366933948,
5547.72357794582,
2442.96833756322,
4535.35536257199,
11068.7231625113,
10676.0333370982,
11455.3286642235,
6842.05778552118,
9439.31658996695,
6966.3414993091,
10999.1589642535,
12438.2086896966,
6791.7839379994,
9862.08653890057,
4306.59412484229,
6975.16764818264,
9980.26897398618,
9312.70761429859,
11279.3297691799,
3174.336,
9235.65661712226,
6444.90877885251,
9436.78767774106,
11519.0999459297,
10207.4488963653,
11491.8371124061,
11931.3766680685,
10039.1662716732,
8351.84656869931,
11844.5646019826,
7700.01994208471,
8499.14824343647,
11444.3585086212,
10659.7688434965,
11939.8919170922,
9799.12857386602,
6272.4515758486,
7126.201929228,
12908.662288495,
7787.12424007209,
11913.2103752478,
9404.56765539201,
5692.32596936017,
10993.155180775,
9330.03591132472,
11105.6148984079,
4222.47887149294,
7673.86524241514,
10543.104,
10496.7546405527,
11668.0546778011,
8400.19000444578,
11625.6989111445,
10079.0619453289,
7634.65065545209,
11433.044589967,
6326.05256881946,
11177.1853380595,
9732.41297446681
)
),
row.names = c(NA, -60L),
class = c("tbl_df", "tbl", "data.frame")
)
Packages
library(tidyverse)
library(emmeans)
library(ggpmisc)
Convert History and Rep to factor
data$History <- factor(data$History)
data$Rep <- data$Rep
Model fitting
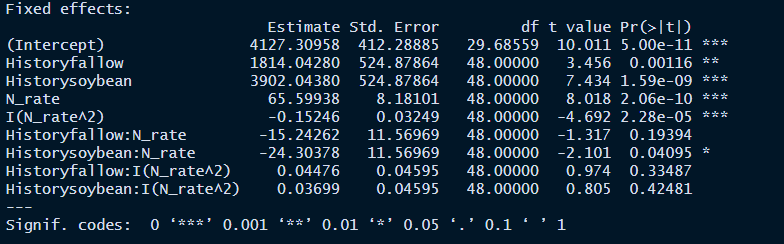
mod3_quad <- lmer(
Yield (lbs/ac) ~ History * N rate + History * I(N rate^2) +
(1 | History:N rate) + (1 | Rep),
data = data
)
Summary
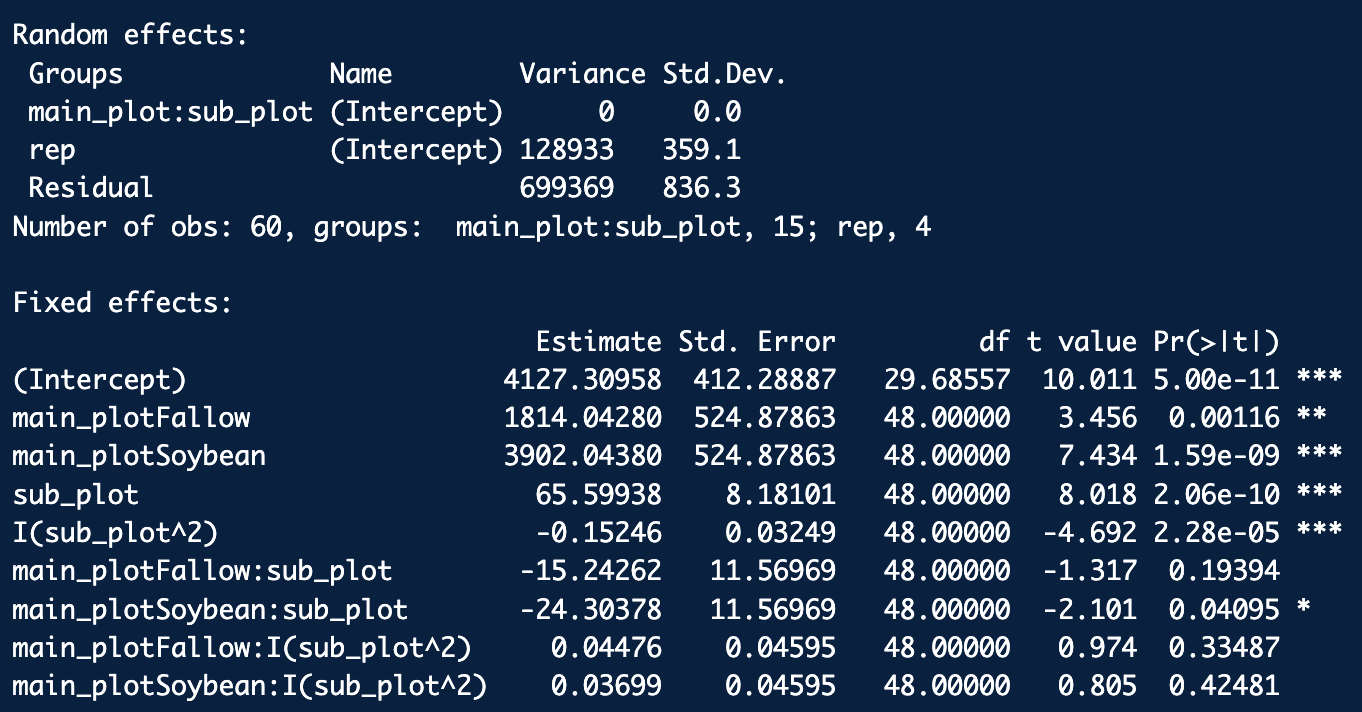
summary(mod3_quad)
anova(mod3_quad)
Post Hoc Test
emmeans::emmeans(object = mod3_quad,specs = ~History) %>% multcomp::cld(Letters=letters,reverse=T)
Augmenting and adding pearson standardized residuals
mod3_quad_aug <- augment(mod3_quad) %>%
mutate(.stdresid = resid(mod3_quad,
type = "pearson",
scaled = T))
mod3_quad_aug
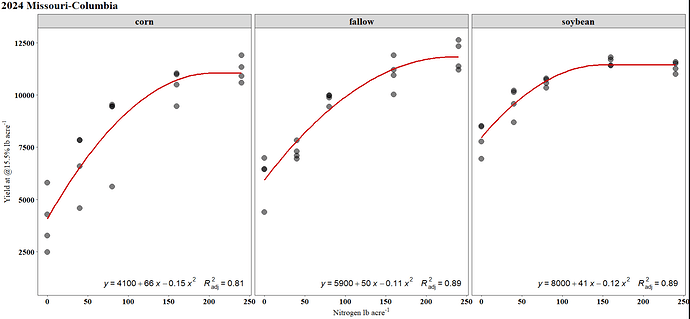
Final Plot
ggplot(mod3_quad_aug,
aes(x = N rate,
y = Yield (lbs/ac)))+
geom_point(size = 3, alpha = .7)+
geom_line(aes(y = .fixed),
color = "red",linewidth=0.7)+
facet_grid(~History)+
labs(y='Yield (lb/ac)',x='N rate (lb/ac)',title = '2024 Missouri-Columbia')+
#stat_poly_line(formula = formula)+
#facet_grid(~History)+
#stat_poly_eq(use_label('R2','P'),label.x = 'right',label.y = 'bottom',size=6)+
#stat_smooth(method = "lm", col = "blue",se=F,formula = y~poly(x,2))+ # This add polynomial curve to the graph
theme_bw()+
coord_cartesian(clip = 'off')+
theme(panel.grid = element_blank(),
strip.text.x = element_text(family = 'serif',face = 'bold',colour = 'black',size = 16),
axis.text = element_text(family = 'serif',face = 'bold',colour = 'black',size = 13),
axis.title = element_text(family = 'serif',face = 'bold',colour = 'black',size = 13),
plot.title = element_text(family = 'serif',face = 'bold',colour = 'black',size = 18),
plot.title.position = 'plot')