Hello RStudio.Community! I’m hoping you can help me with this issue I ran into while creating a custom color scale for ggplot2. I’m sorry for the long-winded post–I followed the excellent posts from Robert Franssen, Simon Jackson, Maddie Pickens, and the well-written documentation for ggplot2 ![]()
![]()
Load packages
Below are the packages I’m using:
library(tidyverse)
library(scales)
library(grDevices)
library(NHANES)
library(janitor)
Colors, palettes, scales
# define colors ----
pckg_colors <- c(
## dark blues ----
`dark_blue_t3` = "#325777",
`dark_blue_t2` = "#194266",
`dark_blue_primary` = "#002E56", # primary
`dark_blue_s2` = "#00294d",
`dark_blue_s3` = "#002444",
## purples ----
`purple_t3` = "#5d63ac",
`purple_t2` = "#4950a2",
`purple_primary` = "#353D98", # primary
`purple_s2` = "#2f3688",
`purple_s3` = "#2a3079",
## blues ----
`blue_t3` = "#3272af",
`blue_t2` = "#1961a5",
`blue_primary` = "#00509C", # primary
`blue_s2` = "#00488c",
`blue_s3` = "#00407c",
## violets ----
`violet_t3` = "#ba4da4",
`violet_t2` = "#b13799",
`violet_primary` = "#A9218E", # primary
`violet_s2` = "#981d7f",
`violet_s3` = "#871a71",
## reds ----
`red_t3` = "#f15971",
`red_t2` = "#ef445f",
`red_primary` = "#EE304E", # primary
`red_s2` = "#d62b46",
`red_s3` = "#be263e",
## oranges ----
`orange_t3` = "#f4845a",
`orange_t2` = "#f37545",
`orange_primary` = "#F26631", # primary
`orange_s2` = "#d95b2c",
`orange_s3` = "#c15127")
# Define function for extracting colors ----
pckg_cols <- function(...) {
cols <- c(...)
if (is.null(cols))
return(pckg_colors)
pckg_colors[cols]
}
# Define list of palettes ----
pckg_palettes <- list(
`primary` = pckg_cols(
"dark_blue_primary", "purple_primary", "blue_primary",
"violet_primary", "red_primary", "orange_primary"
),
`light` = pckg_cols(
"dark_blue_t3", "dark_blue_t2", "purple_t3", "purple_t2",
"blue_t3", "blue_t2", "violet_t3", "violet_t2",
"red_t3", "red_t2", "orange_t3", "orange_t2"
),
`dark` = pckg_cols(
"dark_blue_s2", "dark_blue_s3", "purple_s2", "purple_s3",
"blue_s2", "blue_s3", "violet_s2", "violet_s3",
"red_s2", "red_s3", "orange_s2", "orange_s3"
),
`all` = pckg_cols(
"dark_blue_t3", "dark_blue_t2", "dark_blue_primary",
"dark_blue_s2", "dark_blue_s3",
"purple_t3", "purple_t2", "purple_primary",
"purple_s2", "purple_s3",
"blue_t3", "blue_t2", "blue_primary",
"blue_s2", "blue_s3",
"violet_t3", "violet_t2", "violet_primary",
"violet_s2", "violet_s3",
"red_t3", "red_t2", "red_primary",
"red_s2", "red_s3",
"orange_t3", "orange_t2", "orange_primary",
"orange_s2", "orange_s3"
)
)
# Define function for extracting colors ----
pckg_pal <- function(palette = "main", reverse = FALSE, ...) {
pal <- pckg_palettes[[palette]]
if (reverse) pal <- rev(pal)
grDevices::colorRampPalette(colors = pal, ...)
}
# Define color scale
scale_color_pckg <- function(palette = "main", discrete = TRUE, reverse = FALSE, ...) {
pal <- pckg_pal(palette = palette, reverse = reverse)
if (discrete) {
ggplot2::discrete_scale(aesthetics = "colour",
scale_name = paste0("pckg_", palette), palette = pal, ...)
} else {
ggplot2::scale_color_gradientn(colours = pal(256), ...)
}
}
# Define fill scale
scale_fill_pckg <- function(palette = "main", discrete = TRUE, reverse = FALSE, ...) {
pal <- pckg_pal(palette = palette, reverse = reverse)
if (discrete) {
ggplot2::discrete_scale(aesthetics = "fill",
scale_name = paste0("pckg_", palette), palette = pal, ...)
} else {
ggplot2::scale_fill_gradientn(colours = pal(256), ...)
}
}
Test these below using the nhanes data (a subset of the NHANES data).
nhanes <- NHANES::NHANES %>% janitor::clean_names()
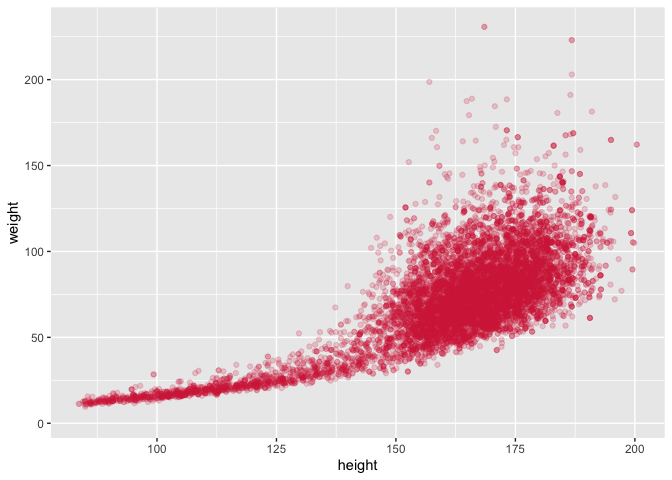
ggplot(data = nhanes,
mapping = aes(x = height, y = weight)) +
geom_point(alpha = 1/5, color = pckg_cols("red_s2"))
## Warning: Removed 366 rows containing missing values (geom_point).
ggplot(data = nhanes,
mapping = aes(x = height, y = weight)) +
geom_point(alpha = 1/5, color = pckg_cols("purple_t2"))
## Warning: Removed 366 rows containing missing values (geom_point).
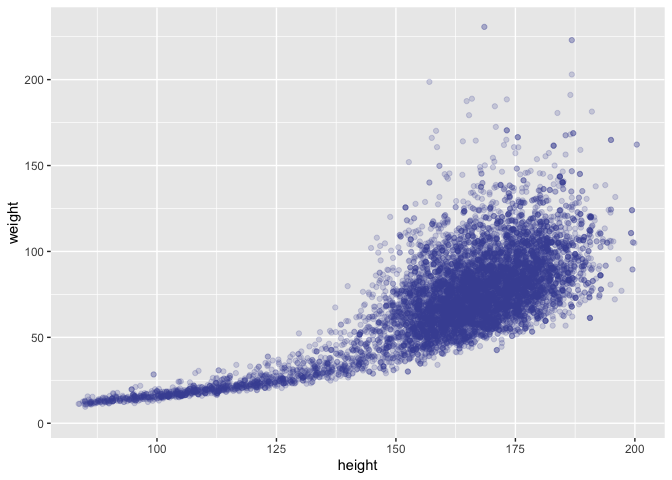
On both of these I see:
## Warning: Removed 366 rows containing missing values (geom_point).
Color aesthetic
ggplot(data = nhanes,
mapping = aes(x = height, y = weight, color = age)) +
geom_point() +
scale_color_pckg(discrete = FALSE, palette = "light")
## Warning: Removed 366 rows containing missing values (geom_point).
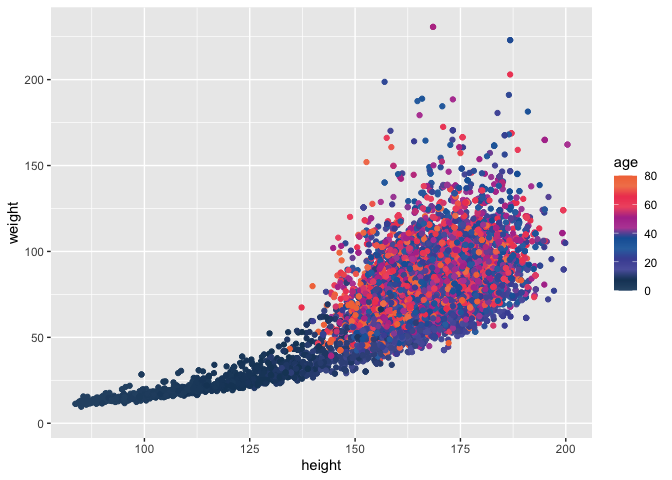
I also see
## Warning: Removed 366 rows containing missing values (geom_point).
Fill aesthetic
When I test the fill aesthetic, I pass it two categorical variables (no missing values).
nhanes %>%
count(race1)
## # A tibble: 5 × 2
## race1 n
## <fct> <int>
## 1 Black 1197
## 2 Hispanic 610
## 3 Mexican 1015
## 4 White 6372
## 5 Other 806
ggplot(data = nhanes,
mapping = aes(x = race1, fill = race1)) +
geom_bar() +
scale_fill_pckg(palette = "light", guide = "none")
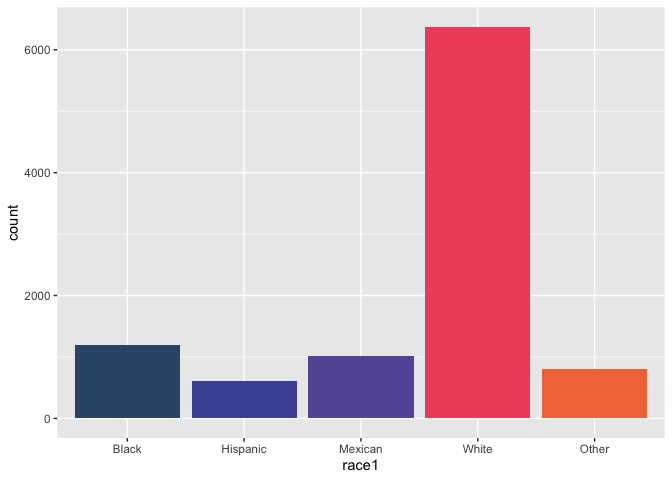
But when I can pass it two variables with missing values…
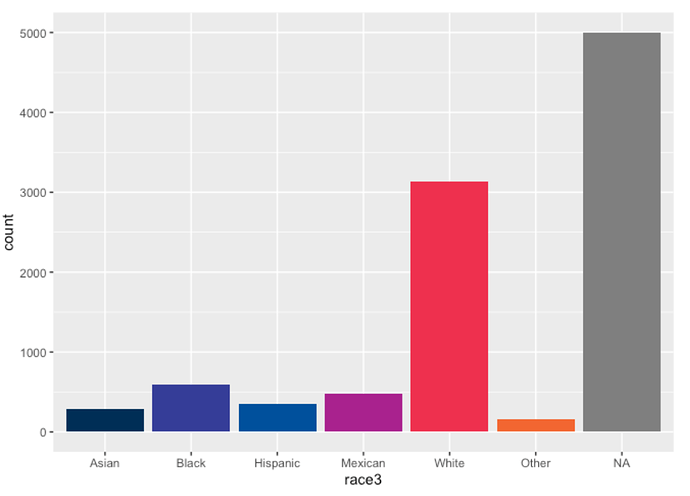
nhanes %>%
count(race3)
## # A tibble: 7 × 2
## race3 n
## <fct> <int>
## 1 Asian 288
## 2 Black 589
## 3 Hispanic 350
## 4 Mexican 480
## 5 White 3135
## 6 Other 158
## 7 <NA> 5000
ggplot(data = nhanes,
mapping = aes(x = race3, fill = race3)) +
geom_bar() +
scale_fill_pckg(palette = "primary", guide = "none")
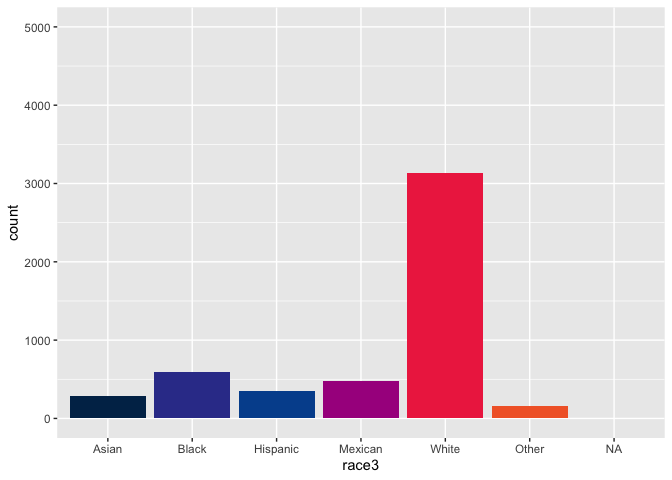
Here I see the missing values for race3 aren’t plotted with this scale? Do I need to provide an argument to na.translate or na.value in the discrete_scale() function?
Any help you can provide would be greatly appreciated! ![]()
![]()