Hi i,m using meta-packages
> glimpse(Book2)
Rows: 4
Columns: 5
$ Author <chr> "A", "B", "C", "D"
$ event.e <dbl> 205, 352, 122, 221
$ n.e <dbl> 221, 352, 125, 222
$ event.c <dbl> 201, 356, 124, 231
$ n.c <dbl> 216, 358, 126, 234
> library(dmetar)
> library(tidyverse)
> library(meta)
> m.bin <- metabin(event.e = event.e,
+ n.e = n.e,
+ event.c = event.c,
+ n.c = n.c,
+ studlab = author,
+ data = Book2,
+ sm = "RR",
+ method = "MH",
+ MH.exact = TRUE,
+ fixed = TRUE,
+ random = TRUE,
+ method.tau = "PM",
+ method.random.ci = "HK",
+ title = "Depression and Mortality")
Error in eval(matchcall[[match(argname, names(matchcall))]], data, enclos = encl) :
object 'author' not found
> m.bin <- metabin(event.e = event.e,
+ n.e = n.e,
+ event.c = event.c,
+ n.c = n.c,
+ studlab = Author,
+ data = Book2,
+ sm = "RR",
+ method = "MH",
+ MH.exact = TRUE,
+ fixed = TRUE,
+ random = TRUE,
+ method.tau = "PM",
+ method.random.ci = "HK",
+ title = "Depression and Mortality")
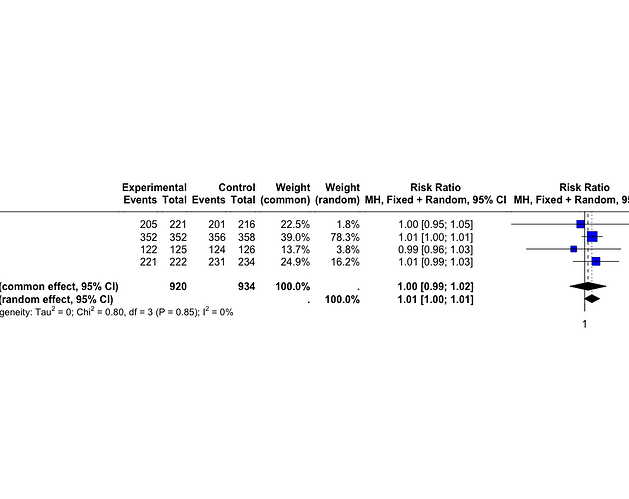
> summary(m.bin)
Review: Depression and Mortality
RR 95%-CI %W(common) %W(random)
A 0.9968 [0.9465; 1.0498] 22.5 1.8
B 1.0056 [0.9978; 1.0134] 39.0 78.3
C 0.9917 [0.9573; 1.0274] 13.7 3.8
D 1.0084 [0.9914; 1.0258] 24.9 16.2
Number of studies: k = 4
Number of observations: o = 1854 (o.e = 920, o.c = 934)
Number of events: e = 1812
RR 95%-CI z|t p-value
Common effect model 1.0024 [0.9889; 1.0162] 0.35 0.7249
Random effects model 1.0054 [0.9996; 1.0112] 2.96 0.0594
Quantifying heterogeneity:
tau^2 = 0 [0.0000; 0.0006]; tau = 0 [0.0000; 0.0251]
I^2 = 0.0% [0.0%; 84.7%]; H = 1.00 [1.00; 2.56]
Test of heterogeneity:
Q d.f. p-value
0.80 3 0.8488
Details on meta-analytical method:
- Mantel-Haenszel method, without continuity correction) (common effect model)
- Inverse variance method (random effects model)
- Paule-Mandel estimator for tau^2
- Q-Profile method for confidence interval of tau^2 and tau
- Hartung-Knapp adjustment for random effects model (df = 3)
- Continuity correction of 0.5 in studies with zero cell frequencies
(only used to calculate individual study results)
> forest(m.bin, layout = "RevMan5")