Hello,
I'm investigating bacterial diversity and want wo know if the diversity is dependent on the pH. My data are structured as followed:
- I have a dataset of 17 different studies
- each study has values for diversity and for pH (the number of values differ between the studies)
Now I am searching for a way to answer the question "Is there a effect of the pH on the diversity throughout all studies"
The idea was to use the function lme and set study as random factor. Looking at the data they seem to rather fit a quadratic term than a linear regression, so I tried to calculate the model also with the quadratic term of the pH:
my_model<- lme( fixed = Bacterial_diversity ~ pH +
I(pH^2),
random = ~ pH |Study,
data= my_data)
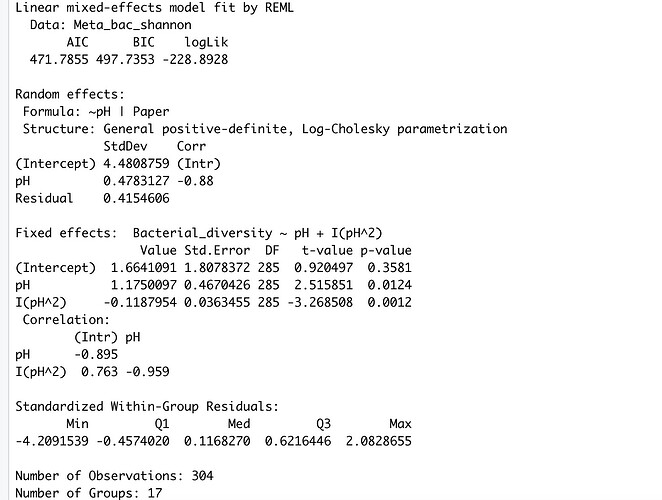
The output (summary) is:
I have absolutely no idea how to interpret these results. When I use tab-model (my_model) I end up with two R^2 values. The marginal R^2 (0.025) and the conditional R^2 (0.974), the given p-values is 0.0012 (for the quadratic term of pH) and 0.0124 (for the pH).
Is the given p-value true for both of the R^2 values or are they calculated for the whole model? and how can I interpret the R^2 values.
Is even the model I used the right one or should I have used nlme instead of lme4 (I don't really understand the difference!).
If you have any ideas this would help a lot.
And if there are questions regarding the data, the code or anything else, please feel free to ask.
Thanks to everyone ![]()