I have my own multiple ELISA(enzyme-linked immunosorbent assay) dataset like following.
> dat
# A tibble: 42 × 3
CURVE conc observation
<fct> <dbl> <dbl>
1 1 0 1.81
2 1 0 1.87
3 1 0 1.96
4 1 0.62 1.39
5 1 0.62 1.16
6 1 0.62 1.06
7 1 1.85 0.994
8 1 1.85 0.833
9 1 1.85 0.833
10 1 5.56 0.725
# … with 32 more rows
I could fit 4 parameters logistic curve to my calibration-standards and get parameters using broom , purrr, drc. (I followed bloom and dplyr, it's pretty efficient way!!)
library(broom)
library(dplyr)
library(tidyr)
library(tibble)
library(ggplot2)
library(purrr)
library(drc)
## Generate calibration standard's data
spinach %>%
select(-HERBICIDE) %>%
rename(conc=DOSE) %>%
rename(observation=SLOPE) %>%
filter(CURVE==1 | CURVE==3) %>%
mutate(CURVE=as.factor(CURVE)) -> dat
## For plotting calibration curve
xx <- seq(0, 150, 0.1)
bind_rows(data.frame(xx=xx, CURVE=1),
data.frame(xx=xx, CURVE=3)) %>%
nest(xx = -CURVE) %>%
mutate(CURVE=as.factor(CURVE)) -> xx
## Regression & drawing curve
dat %>%
nest(data = -CURVE) %>%
left_join(xx, by='CURVE') %>%
mutate(
fit = map(data, ~ drm(observation ~ conc, data = .x, fct=LL.4())),
tidied = map(fit, tidy),
augmented = map2(fit, xx, ~augment(.x, newdata=.y))
)%>%
unnest(augmented, name_repair='unique') %>%
ggplot(aes(x=xx1, y=.fitted, color=CURVE)) +
xlab('conc') +
ylab('observation') +
geom_line() -> g
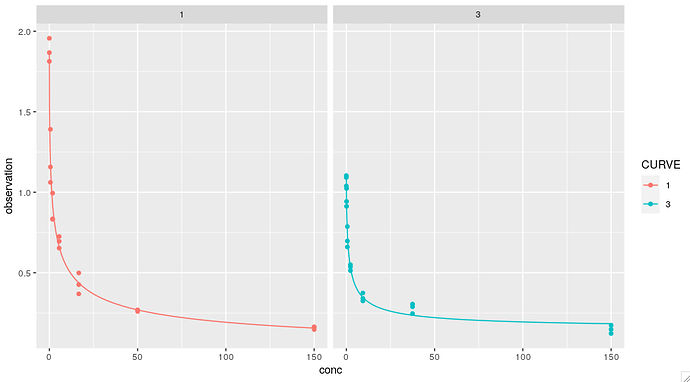
## Add original point
g + geom_point(data=dat, aes(x=conc, y=observation, color=CURVE)) +
facet_wrap(~CURVE) -> g
g
Then, how can I know sample's 'conc' with 'tidy' coding style? I tried to map inverse function of 4PL, but output data type was 'function'(should be 'double').... Thanks!!
## Now I would like to know following unknown sample's conc.. how can I do in efficient way?
new.observation <- bind_rows(data.frame(observation=c(1.6, 0.6, 0.25), CURVE=as.factor(1)),
data.frame(observation=c(0.8, 0.26, 0.2), CURVE=as.factor(3)))
new.observation
## > observation CURVE
## 1 1.60 1
## 2 0.60 1
## 3 0.25 1
## 4 0.80 3
## 5 0.26 3
## 6 0.20 3
inverse.LL4 <- function(fit, observe){
y <- observe[[1]]
b <- fit[[1]]$coefficients[1]
c <- fit[[1]]$coefficients[2]
d <- fit[[1]]$coefficients[3]
e <- fit[[1]]$coefficients[4]
conc <- exp(log((d-y)/(y-c))/b + log(e))
return(conc)
}
new.observation %>%
nest(new.observation= -CURVE) -> no
datd %>%
left_join(no, by='CURVE') %>%
mutate(pred=map2(fit, new.observation, ~inverse.LL4)) %>%
select(pred)
## A tibble: 2 × 1
# pred
# <list>
#1 <fn>
#2 <fn>
#>