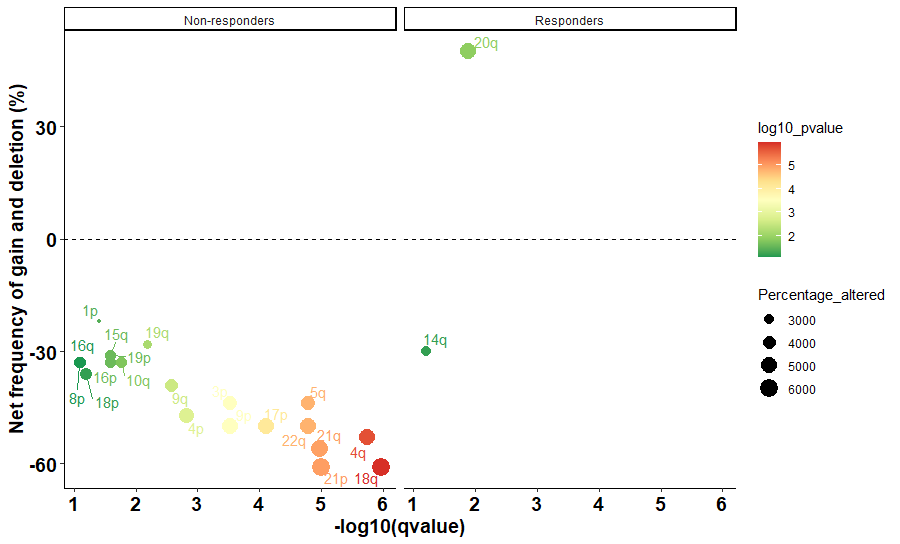
Hello
I have such a data
> dput(x)
structure(list(gene = c("14q", "20q", "18q", "4q", "21p", "21q",
"5q", "22q", "17p", "3p", "9p", "4p", "9q", "19q", "10q", "15q",
"16p", "19p", "1p", "18p", "16q", "8p"), CNV = c("Deletion",
"Amplification", "Deletion", "Deletion", "Deletion", "Deletion",
"Deletion", "Deletion", "Deletion", "Deletion", "Deletion", "Deletion",
"Deletion", "Deletion", "Deletion", "Deletion", "Deletion", "Deletion",
"Deletion", "Deletion", "Deletion", "Deletion"), log10_pvalue = c(1.197226275,
1.88941029, 5.974694135, 5.73754891, 4.995678626, 4.970616222,
4.793174124, 4.793174124, 4.109020403, 3.524328812, 3.524328812,
2.823908741, 2.567030709, 2.186419011, 1.769551079, 1.59345982,
1.59345982, 1.59345982, 1.416801226, 1.195860568, 1.094743951,
1.087777943), Percentage_altered = c(3000, 5000, 6100, 5300,
6100, 5600, 4400, 5000, 5000, 4400, 5000, 4700, 3900, 2800, 3300,
3100, 3300, 3100, 2200, 3600, 3300, 3300), group = c("Responders",
"Responders", "Non-responders", "Non-responders", "Non-responders",
"Non-responders", "Non-responders", "Non-responders", "Non-responders",
"Non-responders", "Non-responders", "Non-responders", "Non-responders",
"Non-responders", "Non-responders", "Non-responders", "Non-responders",
"Non-responders", "Non-responders", "Non-responders", "Non-responders",
"Non-responders")), row.names = c(NA, -22L), class = "data.frame")
I have used this code
> p=x %>%
Warning messages:
1: ggrepel: 6 unlabeled data points (too many overlaps). Consider increasing max.overlaps
2: ggrepel: 6 unlabeled data points (too many overlaps). Consider increasing max.overlaps
+ mutate(net_frequency=ifelse(CNV == "Deletion", -Percentage_altered/100, Percentage_altered/100),
+ log10_pvalue = if_else(CNV == "Deletion", log10_pvalue, log10_pvalue)) %>%
+ ggplot(aes(x = log10_pvalue, y = net_frequency, color = log10_pvalue)) +
+ geom_point(aes(size=Percentage_altered)) +
+ geom_text_repel(aes(label=gene), force=15) +
+ geom_hline(yintercept=0, lty=2) +
+ scale_color_distiller(type = "div",palette = 5) +
+ theme_classic() +
+ facet_wrap(~group)
> p+xlab("-log10(qvalue)")+ylab("Net frequency of gain and deletion (%)")+theme(
+ plot.title = element_text(color="black", size=14, face="bold.italic"),
+ axis.title.x = element_text(color="black", size=14, face="bold"),
+ axis.title.y = element_text(color="black", size=14, face="bold")
+ )+theme(axis.text.x = element_text(face="bold", color="black",
+ size=14),
+ axis.text.y = element_text(face="bold", color="black",
+ size=14))
To plot something like this
You are seeing here, chromosomal arm size like 1p, 14q, whetever are too small too read even color causes some arms are not readable. How I can make the size of these elements bigger please ?
Thank you so much in advance