Hello everyone,
This is my first time posting on a forum so please feel free to give me advice on how to use it. As far as my level on R is concerned, I have a few basics and I'm trying to improve each day.
My problem : I'm trying to create a loop that allows me to output a graph for each column of a table (for now, I can output graphs but they are empty).
Context / Data: When a subject A moves in a square room, I record the position of the subject in the room (XY) and the activity of cells/neurons (between 50 and 200).
I have 2 sets of data:
- The path followed during the experiment (Frame by Frame (one frame=one row of the table), I have access to the X and Y position)
- The activity of cells (between 50 and 200 cells, Frame by Frame for each cell activated; 1 = active and 0 = not active)
What I want:
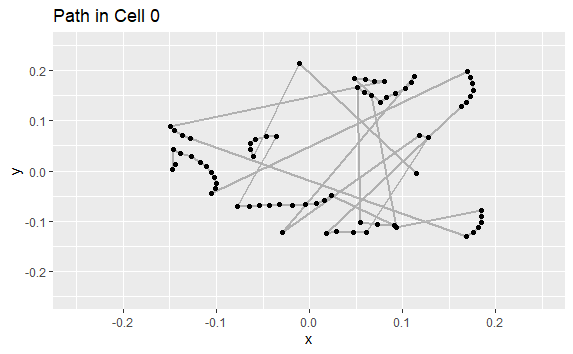
For 1 cell, I want to output an "XY graph that represents the square room" with the path traveled by the subject and overlap the events i.e. place a point when the cell is active (=1)
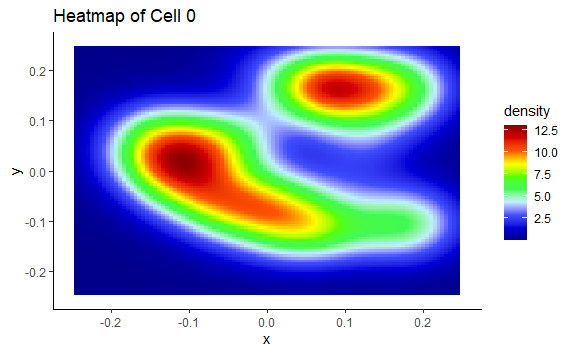
A map of the activity of the cell in the room
=> Given that I have a large number of cells (therefore a large number of columns in my table). I want to create a loop that automatically outputs the graph / activity map of all my cells.
My progress : I have found a way to plot the graphs of interest for a cell. (Here is the code for 1 cells, N°38)
library(ggplot2)
library(grid)
library(dplyr)
library(knitr)
library(data.table)
library(RColorBrewer)
library(reprex)
palette <- colorRampPalette(c("darkblue", "blue", "lightblue1", "green","yellow", "red", "darkred"))
event <- read.csv2("CaDriftT0.csv",header=T,dec = ",") #Cells event
posdrift <- read.csv2("Raw_dataDriftT0.csv",header=T,dec = ",") #Position
x <- posdrift$X.center
y <- posdrift$Y.center
dtall <- data.table(X=x, Y=y, event) #create data table X, Y, Time, time sync, C00, C001, C002, ...
dtall <- select(dtall,-Time.s.,-Time.sync.) #Remove 2 columns : Time.s.,-Time.sync.
suball <- subset(dtall, C38 == "1") #If i change Cells Number, i will have the graph and heatmap for this cell -> ==1 select frame where cell was activated
p <- ggplot(data = suball, aes(X,Y))
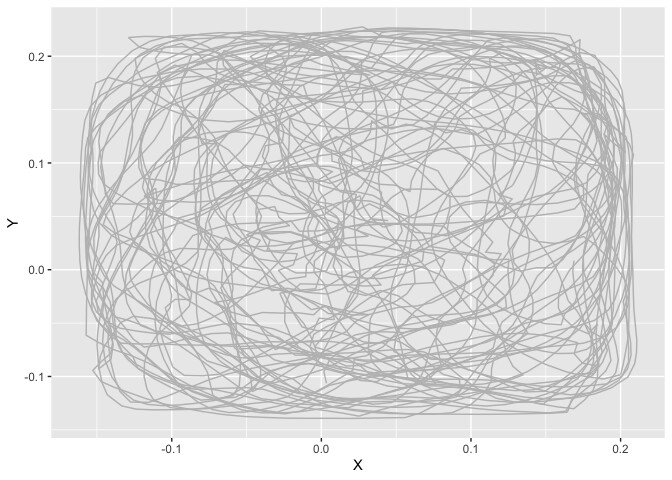
p + geom_path(data = dtall,aes(X,Y),color = "gray")+ geom_point () #See where are the event on track
p + stat_density2d(aes(fill = ..density..), geom = "tile", contour = FALSE, n = 32) + scale_fill_gradientn(colors = palette(10))+ theme_classic() #Heatmap
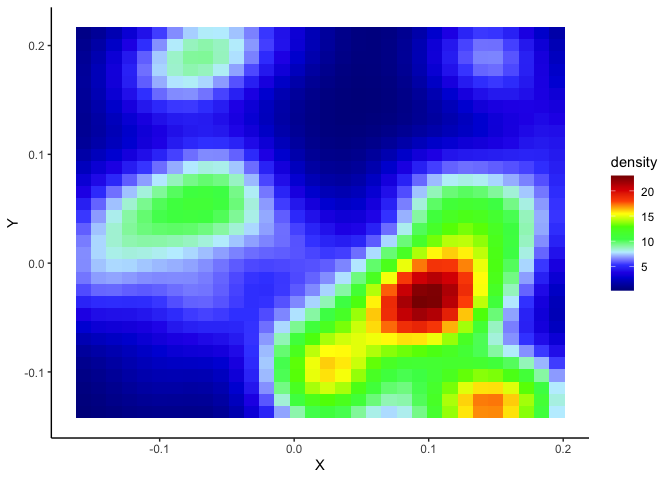
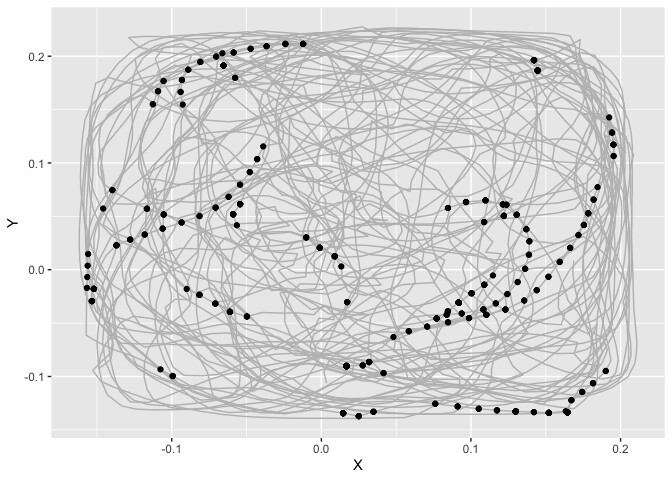
I have started to create a loop that repeats the main steps (starting with column 60 to reduce the size of the compilation file). In this loop, my logic is that if I change the column/cell specified in the subset() command, then I change the column/cell at each turn of the loop and the data taken into account in the graphs.
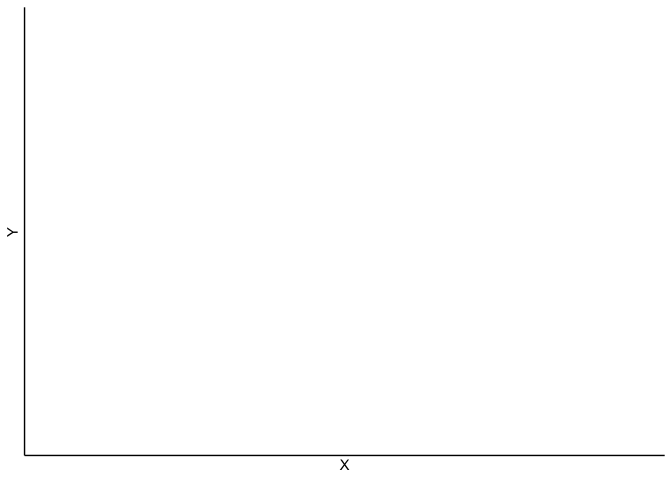
But, when I compiled the document to see what I get: I do get the plot via the X and Y positions of the data.table "dtall", however, the points representing the "1" events in the cell do not appear. Similarly for the heatmap of events, it only creates an empty graph (with the XY axis)
## Loop ## -> I chose 70 at random in order to reduce the number of columns to process
for (i in 70:ncol(dtall)){
suballo = subset(dtall, i == "1")
p <- ggplot(data = suballo, aes(X,Y))
print(p + geom_path(data = dtall,aes(X,Y),color = "gray")+ geom_point ())
print(p + stat_density2d(aes(fill = ..density..), geom = "tile", contour = FALSE, n = 16) + scale_fill_gradientn(colors = palette(10))+ theme_classic())
}
I am sharing the .csv files on this Github to visualize the data, which I hope is a safe way for you ! GitHub - Anthorhinal/FilesRstudioForum: Csv files
Other information : I don't use data.frame because the length of the columns are different. That's why I chose data.table. I hope this does not cause any problems. Also, I was thinking about the subset()as a problem in the loop.
I thank you in advance for the help you will bring me.
Anthorhinal,