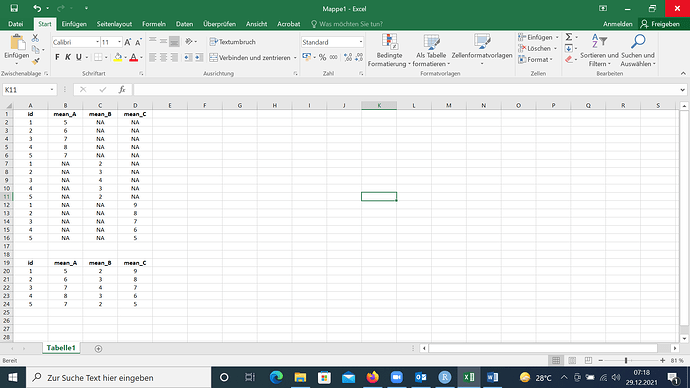
The data frame isn't really in long format, but it's not properly combined. If there are previous steps in the data shaping, it's possible you can avoid the problem by revising the earlier code. In any case, given the current data frame, you could do the following: In the code below, we keep only the non-missing values from each column and then bind that back to a single copy of the IDs
library(tidyverse)
# Fake data
d = tibble(id=rep(1:5, 3),
mean_a=c(1:5,rep(NA,10)),
mean_b=c(rep(NA,5),6:10,rep(NA,5)),
mean_c=c(rep(NA,10),11:15))
distinct(d, id) %>%
bind_cols(
d %>%
select(-id) %>%
map_df(na.omit)
)
#> # A tibble: 5 × 4
#> id mean_a mean_b mean_c
#> <int> <int> <int> <int>
#> 1 1 1 6 11
#> 2 2 2 7 12
#> 3 3 3 8 13
#> 4 4 4 9 14
#> 5 5 5 10 15
It might actually be safer to use a join, so that we don't have to count on the data being in the right positions to match properly. For example:
library(tidyverse)
# Fake data
d = tibble(id=rep(1:5, 3),
mean_a=c(1:5,rep(NA,10)),
mean_b=c(rep(NA,5),6:10,rep(NA,5)),
mean_c=c(rep(NA,10),11:15))
paste0("mean_", letters[1:3]) %>%
map(
~d %>%
select(id, all_of(.x)) %>%
na.omit
) %>%
reduce(left_join)
#> Joining, by = "id"
#> Joining, by = "id"
#> # A tibble: 5 × 4
#> id mean_a mean_b mean_c
#> <int> <int> <int> <int>
#> 1 1 1 6 11
#> 2 2 2 7 12
#> 3 3 3 8 13
#> 4 4 4 9 14
#> 5 5 5 10 15