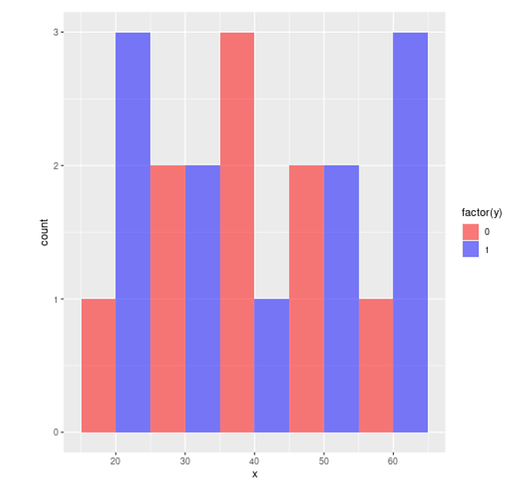
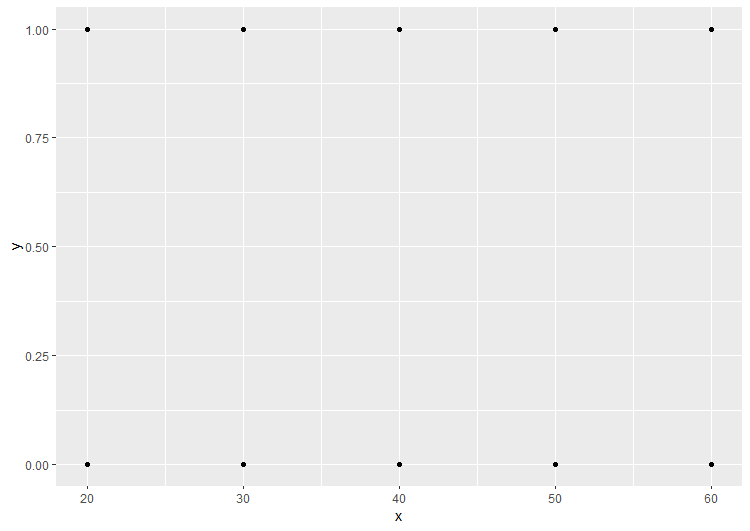
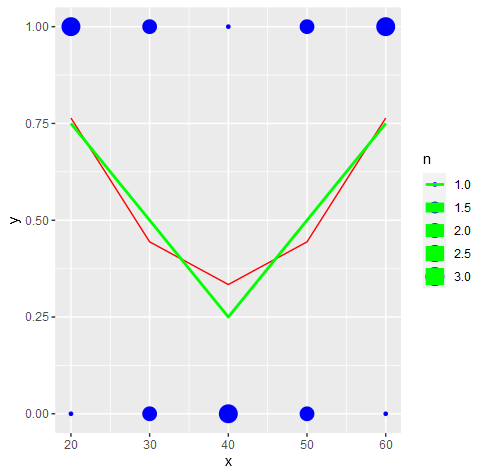
If x is continuous it's hard to see where any glm model will go
# Create the data frame
d <- data.frame(x = c(20,30,30,40,40,40,50,50,60, 20,20,20,30,30,40,50,50,60,60,60),
y = c(0,0,0,0,0,0,0,0,0,1,1,1,1,1,1,1,1,1,1,1))
# Define the formulas and family arguments
formulas <- list(y ~ x, y ~ I(x^2), y ~ I(sin(x)), y ~ I(sin(x^2)))
families <- list(binomial(link = "logit"),
gaussian(link = "identity"),
poisson(link = "log"),
quasi(link = "identity", variance = "constant"),
quasibinomial(link = "logit"),
quasipoisson(link = "log"))
# Fit the models and store them in a list
models <- list()
for (f in formulas) {
for (family in families) {
model <- glm(f, data = d, family = family)
models <- append(models, list(model))
}
}
lapply(models,summary)
#> [[1]]
#>
#> Call:
#> glm(formula = f, family = family, data = d)
#>
#> Coefficients:
#> Estimate Std. Error z value Pr(>|z|)
#> (Intercept) 2.007e-01 1.348e+00 0.149 0.882
#> x 1.600e-18 3.178e-02 0.000 1.000
#>
#> (Dispersion parameter for binomial family taken to be 1)
#>
#> Null deviance: 27.526 on 19 degrees of freedom
#> Residual deviance: 27.526 on 18 degrees of freedom
#> AIC: 31.526
#>
#> Number of Fisher Scoring iterations: 3
#>
#>
#> [[2]]
#>
#> Call:
#> glm(formula = f, family = family, data = d)
#>
#> Coefficients:
#> Estimate Std. Error t value Pr(>|t|)
#> (Intercept) 5.500e-01 3.518e-01 1.563 0.135
#> x 3.806e-19 8.292e-03 0.000 1.000
#>
#> (Dispersion parameter for gaussian family taken to be 0.275)
#>
#> Null deviance: 4.95 on 19 degrees of freedom
#> Residual deviance: 4.95 on 18 degrees of freedom
#> AIC: 34.831
#>
#> Number of Fisher Scoring iterations: 2
#>
#>
#> [[3]]
#>
#> Call:
#> glm(formula = f, family = family, data = d)
#>
#> Coefficients:
#> Estimate Std. Error z value Pr(>|z|)
#> (Intercept) -5.978e-01 9.045e-01 -0.661 0.509
#> x 6.096e-18 2.132e-02 0.000 1.000
#>
#> (Dispersion parameter for poisson family taken to be 1)
#>
#> Null deviance: 13.152 on 19 degrees of freedom
#> Residual deviance: 13.152 on 18 degrees of freedom
#> AIC: 39.152
#>
#> Number of Fisher Scoring iterations: 5
#>
#>
#> [[4]]
#>
#> Call:
#> glm(formula = f, family = family, data = d)
#>
#> Coefficients:
#> Estimate Std. Error t value Pr(>|t|)
#> (Intercept) 5.500e-01 3.518e-01 1.563 0.135
#> x 3.806e-19 8.292e-03 0.000 1.000
#>
#> (Dispersion parameter for quasi family taken to be 0.275)
#>
#> Null deviance: 4.95 on 19 degrees of freedom
#> Residual deviance: 4.95 on 18 degrees of freedom
#> AIC: NA
#>
#> Number of Fisher Scoring iterations: 2
#>
#>
#> [[5]]
#>
#> Call:
#> glm(formula = f, family = family, data = d)
#>
#> Coefficients:
#> Estimate Std. Error t value Pr(>|t|)
#> (Intercept) 2.007e-01 1.421e+00 0.141 0.889
#> x 1.600e-18 3.350e-02 0.000 1.000
#>
#> (Dispersion parameter for quasibinomial family taken to be 1.111123)
#>
#> Null deviance: 27.526 on 19 degrees of freedom
#> Residual deviance: 27.526 on 18 degrees of freedom
#> AIC: NA
#>
#> Number of Fisher Scoring iterations: 3
#>
#>
#> [[6]]
#>
#> Call:
#> glm(formula = f, family = family, data = d)
#>
#> Coefficients:
#> Estimate Std. Error t value Pr(>|t|)
#> (Intercept) -5.978e-01 6.396e-01 -0.935 0.362
#> x 6.096e-18 1.508e-02 0.000 1.000
#>
#> (Dispersion parameter for quasipoisson family taken to be 0.5000009)
#>
#> Null deviance: 13.152 on 19 degrees of freedom
#> Residual deviance: 13.152 on 18 degrees of freedom
#> AIC: NA
#>
#> Number of Fisher Scoring iterations: 5
#>
#>
#> [[7]]
#>
#> Call:
#> glm(formula = f, family = family, data = d)
#>
#> Coefficients:
#> Estimate Std. Error z value Pr(>|z|)
#> (Intercept) 0.0338639 0.8366883 0.040 0.968
#> I(x^2) 0.0000930 0.0003948 0.236 0.814
#>
#> (Dispersion parameter for binomial family taken to be 1)
#>
#> Null deviance: 27.526 on 19 degrees of freedom
#> Residual deviance: 27.470 on 18 degrees of freedom
#> AIC: 31.47
#>
#> Number of Fisher Scoring iterations: 4
#>
#>
#> [[8]]
#>
#> Call:
#> glm(formula = f, family = family, data = d)
#>
#> Coefficients:
#> Estimate Std. Error t value Pr(>|t|)
#> (Intercept) 5.087e-01 2.183e-01 2.330 0.0317 *
#> I(x^2) 2.294e-05 1.024e-04 0.224 0.8253
#> ---
#> Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
#>
#> (Dispersion parameter for gaussian family taken to be 0.2742355)
#>
#> Null deviance: 4.9500 on 19 degrees of freedom
#> Residual deviance: 4.9362 on 18 degrees of freedom
#> AIC: 34.775
#>
#> Number of Fisher Scoring iterations: 2
#>
#>
#> [[9]]
#>
#> Call:
#> glm(formula = f, family = family, data = d)
#>
#> Coefficients:
#> Estimate Std. Error z value Pr(>|z|)
#> (Intercept) -6.734e-01 5.712e-01 -1.179 0.238
#> I(x^2) 4.137e-05 2.616e-04 0.158 0.874
#>
#> (Dispersion parameter for poisson family taken to be 1)
#>
#> Null deviance: 13.152 on 19 degrees of freedom
#> Residual deviance: 13.128 on 18 degrees of freedom
#> AIC: 39.128
#>
#> Number of Fisher Scoring iterations: 5
#>
#>
#> [[10]]
#>
#> Call:
#> glm(formula = f, family = family, data = d)
#>
#> Coefficients:
#> Estimate Std. Error t value Pr(>|t|)
#> (Intercept) 5.087e-01 2.183e-01 2.330 0.0317 *
#> I(x^2) 2.294e-05 1.024e-04 0.224 0.8253
#> ---
#> Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
#>
#> (Dispersion parameter for quasi family taken to be 0.2742355)
#>
#> Null deviance: 4.9500 on 19 degrees of freedom
#> Residual deviance: 4.9362 on 18 degrees of freedom
#> AIC: NA
#>
#> Number of Fisher Scoring iterations: 2
#>
#>
#> [[11]]
#>
#> Call:
#> glm(formula = f, family = family, data = d)
#>
#> Coefficients:
#> Estimate Std. Error t value Pr(>|t|)
#> (Intercept) 0.033864 0.881800 0.038 0.970
#> I(x^2) 0.000093 0.000416 0.224 0.826
#>
#> (Dispersion parameter for quasibinomial family taken to be 1.110741)
#>
#> Null deviance: 27.526 on 19 degrees of freedom
#> Residual deviance: 27.470 on 18 degrees of freedom
#> AIC: NA
#>
#> Number of Fisher Scoring iterations: 4
#>
#>
#> [[12]]
#>
#> Call:
#> glm(formula = f, family = family, data = d)
#>
#> Coefficients:
#> Estimate Std. Error t value Pr(>|t|)
#> (Intercept) -6.734e-01 4.040e-01 -1.667 0.113
#> I(x^2) 4.137e-05 1.850e-04 0.224 0.826
#>
#> (Dispersion parameter for quasipoisson family taken to be 0.5003108)
#>
#> Null deviance: 13.152 on 19 degrees of freedom
#> Residual deviance: 13.128 on 18 degrees of freedom
#> AIC: NA
#>
#> Number of Fisher Scoring iterations: 5
#>
#>
#> [[13]]
#>
#> Call:
#> glm(formula = f, family = family, data = d)
#>
#> Coefficients:
#> Estimate Std. Error z value Pr(>|z|)
#> (Intercept) 0.20207 0.44983 0.449 0.653
#> I(sin(x)) -0.06305 0.63278 -0.100 0.921
#>
#> (Dispersion parameter for binomial family taken to be 1)
#>
#> Null deviance: 27.526 on 19 degrees of freedom
#> Residual deviance: 27.516 on 18 degrees of freedom
#> AIC: 31.516
#>
#> Number of Fisher Scoring iterations: 3
#>
#>
#> [[14]]
#>
#> Call:
#> glm(formula = f, family = family, data = d)
#>
#> Coefficients:
#> Estimate Std. Error t value Pr(>|t|)
#> (Intercept) 0.5503 0.1173 4.692 0.000181 ***
#> I(sin(x)) -0.0156 0.1650 -0.095 0.925705
#> ---
#> Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
#>
#> (Dispersion parameter for gaussian family taken to be 0.2748634)
#>
#> Null deviance: 4.9500 on 19 degrees of freedom
#> Residual deviance: 4.9475 on 18 degrees of freedom
#> AIC: 34.821
#>
#> Number of Fisher Scoring iterations: 2
#>
#>
#> [[15]]
#>
#> Call:
#> glm(formula = f, family = family, data = d)
#>
#> Coefficients:
#> Estimate Std. Error z value Pr(>|z|)
#> (Intercept) -0.59746 0.30152 -1.981 0.0475 *
#> I(sin(x)) -0.02837 0.42441 -0.067 0.9467
#> ---
#> Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
#>
#> (Dispersion parameter for poisson family taken to be 1)
#>
#> Null deviance: 13.152 on 19 degrees of freedom
#> Residual deviance: 13.148 on 18 degrees of freedom
#> AIC: 39.148
#>
#> Number of Fisher Scoring iterations: 5
#>
#>
#> [[16]]
#>
#> Call:
#> glm(formula = f, family = family, data = d)
#>
#> Coefficients:
#> Estimate Std. Error t value Pr(>|t|)
#> (Intercept) 0.5503 0.1173 4.692 0.000181 ***
#> I(sin(x)) -0.0156 0.1650 -0.095 0.925705
#> ---
#> Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
#>
#> (Dispersion parameter for quasi family taken to be 0.2748634)
#>
#> Null deviance: 4.9500 on 19 degrees of freedom
#> Residual deviance: 4.9475 on 18 degrees of freedom
#> AIC: NA
#>
#> Number of Fisher Scoring iterations: 2
#>
#>
#> [[17]]
#>
#> Call:
#> glm(formula = f, family = family, data = d)
#>
#> Coefficients:
#> Estimate Std. Error t value Pr(>|t|)
#> (Intercept) 0.20207 0.47416 0.426 0.675
#> I(sin(x)) -0.06305 0.66702 -0.095 0.926
#>
#> (Dispersion parameter for quasibinomial family taken to be 1.111133)
#>
#> Null deviance: 27.526 on 19 degrees of freedom
#> Residual deviance: 27.516 on 18 degrees of freedom
#> AIC: NA
#>
#> Number of Fisher Scoring iterations: 3
#>
#>
#> [[18]]
#>
#> Call:
#> glm(formula = f, family = family, data = d)
#>
#> Coefficients:
#> Estimate Std. Error t value Pr(>|t|)
#> (Intercept) -0.59746 0.21321 -2.802 0.0118 *
#> I(sin(x)) -0.02837 0.30010 -0.095 0.9257
#> ---
#> Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
#>
#> (Dispersion parameter for quasipoisson family taken to be 0.4999959)
#>
#> Null deviance: 13.152 on 19 degrees of freedom
#> Residual deviance: 13.148 on 18 degrees of freedom
#> AIC: NA
#>
#> Number of Fisher Scoring iterations: 5
#>
#>
#> [[19]]
#>
#> Call:
#> glm(formula = f, family = family, data = d)
#>
#> Coefficients:
#> Estimate Std. Error z value Pr(>|z|)
#> (Intercept) 0.2008760 0.4939680 0.407 0.684
#> I(sin(x^2)) 0.0006551 0.6539372 0.001 0.999
#>
#> (Dispersion parameter for binomial family taken to be 1)
#>
#> Null deviance: 27.526 on 19 degrees of freedom
#> Residual deviance: 27.526 on 18 degrees of freedom
#> AIC: 31.526
#>
#> Number of Fisher Scoring iterations: 3
#>
#>
#> [[20]]
#>
#> Call:
#> glm(formula = f, family = family, data = d)
#>
#> Coefficients:
#> Estimate Std. Error t value Pr(>|t|)
#> (Intercept) 0.5500508 0.1288682 4.268 0.000462 ***
#> I(sin(x^2)) 0.0001621 0.1706006 0.001 0.999252
#> ---
#> Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
#>
#> (Dispersion parameter for gaussian family taken to be 0.275)
#>
#> Null deviance: 4.95 on 19 degrees of freedom
#> Residual deviance: 4.95 on 18 degrees of freedom
#> AIC: 34.831
#>
#> Number of Fisher Scoring iterations: 2
#>
#>
#> [[21]]
#>
#> Call:
#> glm(formula = f, family = family, data = d)
#>
#> Coefficients:
#> Estimate Std. Error z value Pr(>|z|)
#> (Intercept) -0.5977447 0.3313258 -1.804 0.0712 .
#> I(sin(x^2)) 0.0002948 0.4386113 0.001 0.9995
#> ---
#> Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
#>
#> (Dispersion parameter for poisson family taken to be 1)
#>
#> Null deviance: 13.152 on 19 degrees of freedom
#> Residual deviance: 13.152 on 18 degrees of freedom
#> AIC: 39.152
#>
#> Number of Fisher Scoring iterations: 5
#>
#>
#> [[22]]
#>
#> Call:
#> glm(formula = f, family = family, data = d)
#>
#> Coefficients:
#> Estimate Std. Error t value Pr(>|t|)
#> (Intercept) 0.5500508 0.1288682 4.268 0.000462 ***
#> I(sin(x^2)) 0.0001621 0.1706006 0.001 0.999252
#> ---
#> Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
#>
#> (Dispersion parameter for quasi family taken to be 0.275)
#>
#> Null deviance: 4.95 on 19 degrees of freedom
#> Residual deviance: 4.95 on 18 degrees of freedom
#> AIC: NA
#>
#> Number of Fisher Scoring iterations: 2
#>
#>
#> [[23]]
#>
#> Call:
#> glm(formula = f, family = family, data = d)
#>
#> Coefficients:
#> Estimate Std. Error t value Pr(>|t|)
#> (Intercept) 0.2008760 0.5206907 0.386 0.704
#> I(sin(x^2)) 0.0006551 0.6893139 0.001 0.999
#>
#> (Dispersion parameter for quasibinomial family taken to be 1.111123)
#>
#> Null deviance: 27.526 on 19 degrees of freedom
#> Residual deviance: 27.526 on 18 degrees of freedom
#> AIC: NA
#>
#> Number of Fisher Scoring iterations: 3
#>
#>
#> [[24]]
#>
#> Call:
#> glm(formula = f, family = family, data = d)
#>
#> Coefficients:
#> Estimate Std. Error t value Pr(>|t|)
#> (Intercept) -0.5977447 0.2342830 -2.551 0.020 *
#> I(sin(x^2)) 0.0002948 0.3101453 0.001 0.999
#> ---
#> Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
#>
#> (Dispersion parameter for quasipoisson family taken to be 0.5000009)
#>
#> Null deviance: 13.152 on 19 degrees of freedom
#> Residual deviance: 13.152 on 18 degrees of freedom
#> AIC: NA
#>
#> Number of Fisher Scoring iterations: 5
Created on 2023-07-09 with reprex v2.0.2