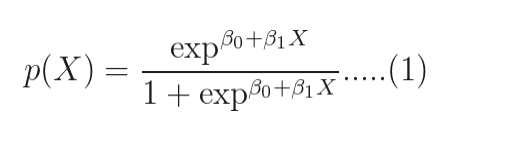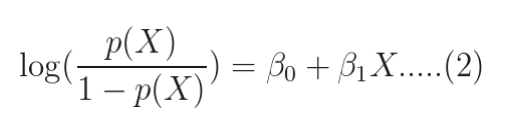@JohnMount
Hello John,
Question 1:
> glm_model <- glm(down_train$Class~.,data = down_train, family = binomial)
Warning message:
glm.fit: fitted probabilities numerically 0 or 1 occurred
> summary(glm_model)
Call:
glm(formula = down_train$Class ~ ., family = binomial, data = down_train)
Deviance Residuals:
Min 1Q Median 3Q Max
-2.1399 -1.0367 0.2300 0.9208 8.4904
Coefficients:
Estimate Std. Error z value Pr(>|z|)
(Intercept) -11.4108728 23.4467343 -0.487 0.62649
cust_prog_levelC 1.1169289 0.1379501 8.097 0.000000000000000565 ***
cust_prog_levelD 0.9901193 0.1298182 7.627 0.000000000000024034 ***
cust_prog_levelE 1.1544305 0.1338535 8.625 < 0.0000000000000002 ***
cust_prog_levelG 0.9318156 0.1293693 7.203 0.000000000000590075 ***
cust_prog_levelI 1.2254934 0.1324209 9.255 < 0.0000000000000002 ***
cust_prog_levelL 1.2905412 0.1296241 9.956 < 0.0000000000000002 ***
cust_prog_levelM 0.9916104 0.3064315 3.236 0.00121 **
cust_prog_levelN 1.2756395 0.1293191 9.864 < 0.0000000000000002 ***
cust_prog_levelP 0.9498830 0.1293275 7.345 0.000000000000206089 ***
cust_prog_levelR 0.7728894 0.2732953 2.828 0.00468 **
cust_prog_levelS 0.9850813 0.1294868 7.608 0.000000000000027928 ***
cust_prog_levelX 1.7231817 0.3874403 4.448 0.000008683257380494 ***
cust_prog_levelZ 2.2222995 0.1342246 16.557 < 0.0000000000000002 ***
CUST_REGION_DESCRMOUNTAIN WEST REGION 11.0671194 23.4463814 0.472 0.63691
CUST_REGION_DESCRNORTH CENTRAL REGION 11.2638726 23.4463804 0.480 0.63094
CUST_REGION_DESCRNORTH EAST REGION 11.3171832 23.4463806 0.483 0.62932
CUST_REGION_DESCROHIO VALLEY REGION 11.3389408 23.4463811 0.484 0.62866
CUST_REGION_DESCRSOUTH CENTRAL REGION 11.2946499 23.4463802 0.482 0.63000
CUST_REGION_DESCRSOUTH EAST REGION 11.3990264 23.4463799 0.486 0.62684
CUST_REGION_DESCRWESTERN REGION 11.1147124 23.4463816 0.474 0.63547
Sales -0.0107124 0.0000521 -205.598 < 0.0000000000000002 ***
---
Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
(Dispersion parameter for binomial family taken to be 1)
Null deviance: 522966 on 377239 degrees of freedom
Residual deviance: 431903 on 377218 degrees of freedom
AIC: 431947
Number of Fisher Scoring iterations: 9
So the summary() give me this result and there are quite a few estimates with standard errors exceed 23. So can I assume the warning message "glm.fit: fitted probabilities numerically 0 or 1 occurred" explains for this large number? I am asking this question because the other standard errors are small and those mentioned above are so so large, so I am curious why!
Question 2: I took your advice and used glmnet() method
>xfactors<-model.matrix(Class ~ CUST_REGION_DESCR + cust_prog_level,data=down_train)[,-1]
>x = as.matrix(data.frame(down_train$Sales,xfactors))
>glmmod = glmnet(x,y=as.factor(down_train$Class),alpha=1,family='binomial')
> summary(glmmod)
Length Class Mode
a0 60 -none- numeric
beta 1260 dgCMatrix S4
df 60 -none- numeric
dim 2 -none- numeric
lambda 60 -none- numeric
dev.ratio 60 -none- numeric
nulldev 1 -none- numeric
npasses 1 -none- numeric
jerr 1 -none- numeric
offset 1 -none- logical
classnames 2 -none- character
call 5 -none- call
nobs 1 -none- numeric
What does the result tell me?
One article says that:
if you take logarithm on both side of the below equation,
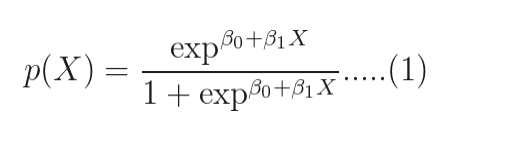
you will get:
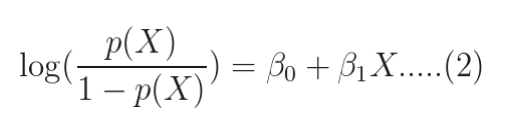
The quantity on the left is the logarithm of the odds. So, the model is a linear regression of the log-odds, sometimes called logit, and hence the name logistic.
Since I have more than one regressor(Sales, cust_program_level, CUST_REGION_DESCR), I assume I will have beta_0, beta_1, beta_2 or something like that.
Should the explanation for summary(glmmod) be focused on whether we have some estimates for those betas just as summary() for a linear regression model?
Thank you!